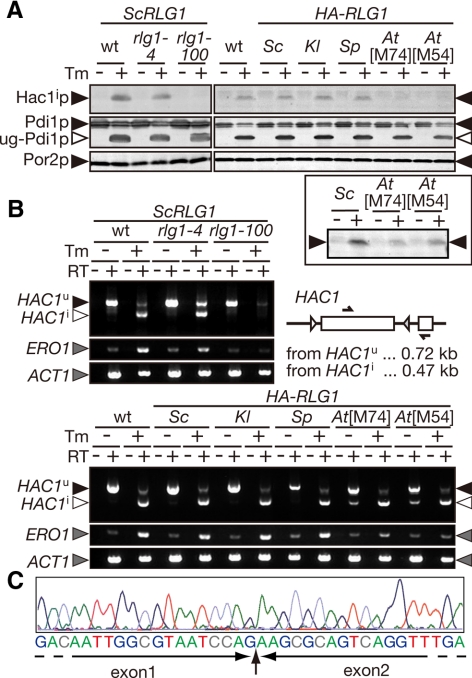
Figure 2.
AtRlg1 proteins can splice HAC1 mRNA, whereas AtRLG1 strains are defective in the UPR. (A) Lysates were prepared from cultures of indicated strains in the absence (−) or presence (+) of 2.0 μg/ml Tm and then analyzed by Western blotting with anti-Hac1p antibodies (top). Appearance of unglycosylated Pdi1p (◁, ug-Pdi1p) was monitored to confirm effects of Tm (middle), and Por2p (mitochondrial porin) was used as a loading control (bottom). In the left part, yeast strains harboring ScRLG1 genes with no mutation (wt), rlg1-4 mutation (defective only in tRNA ligation), or rlg1-100 mutation (defective only in HAC1 mRNA ligation) on their chromosome were analyzed as control. Right, lysates from rlg1Δ strains complemented with various RLG1 genes were analyzed. In the bottom square, twofold more of lysates and the fivefold higher concentration of the anti-Hac1p antibodies were used to detect small amounts of Hac1p produced in the AtRLG1[M74] and AtRLG1[M54] cells. (B) Total RNAs (0.10 μg) prepared from the same set of strains in A was subjected to RT-PCR of HAC1 with a set of primers whose target was represented in the schematic drawing (arrows). ERO1 RT-PCR was also carried out to monitor the UPR, and ACT1 RT-PCR was done as a loading control. The top part represents RT-PCR products from yeast cells with ScRLG1 alleles on their chromosome. In the bottom panel, RT-PCR products from rlg1Δ strains complemented with various RLG1 genes on plasmids were analyzed. RT, reverse-transcription. ▶, RT-PCR fragment derived from HAC1u mRNA; ◁, that from HAC1i mRNA; gray triangles, RT-PCR fragments of ERO1 and ACT1 mRNAs. (C) A typical electrogram of sequencing reactions of the RT-PCR fragment amplified from the AtRLG1[M74] cells treated with Tm. Corresponding sequence is shown beneath the electrogram. The exon–exon junction is indicated by a vertical arrow.