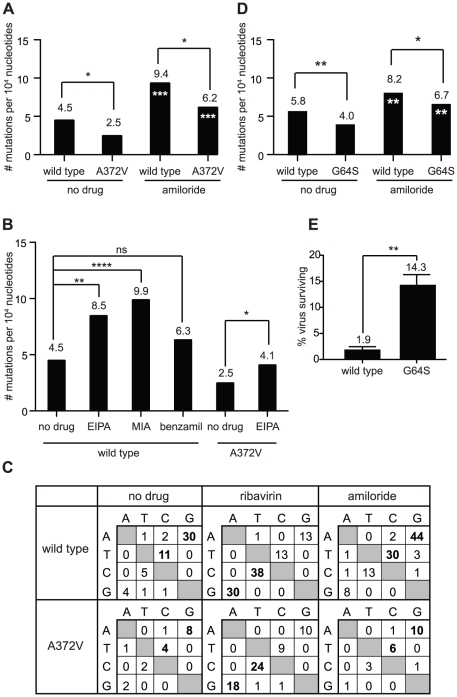
Figure 4. Amiloride has RNA mutagenic activity to which high fidelity RdRp variants of picornaviruses resist.
(A) The mutation frequencies of wild type and A372V viruses grown in the presence of 400 µM amiloride were determined, shown as the mean number of mutations per 104 nucleotides sequenced. * P<0.05 statistically significant difference between WT and A372V. Asterisks in white indicate statistically significant difference between amiloride-treated virus and the untreated parental population, *** P<0.001. (B) Mutations frequencies of wild type and A372V viruses in the presence of other amiloride compounds. HeLa cells were treated with 40 µM of EIPA, 25 µM of MIA or 20 µM of benzamil and were infected with wild type CVB3 virus or A372V variant (EIPA only). Statistical significance of differences in mutation frequencies are indicated. ns = not significant, * P<0.05, ** P<0.005, **** P<0.0001. (C) Mutation profiles of wild type and A372V populations grown in the absence of drug or presence of 400 µm ribavirin or amiloride in (A). The most commonly occurring mutations are indicated in bold. (D) Mutation frequency of wild type and G64S polioviruses. Viral RNA genomes were extracted following infection of HeLa cells, grown in standard conditions or treated with 400 µM amiloride. A 1.0 kb region of the viral capsid was RT-PCR amplified and subcloned for sequencing of individual clones to obtain the observed mutation frequencies (Table S1). Asterisks in white indicate statistical significance between drug treated populations and the same untreated parental virus. * P<0.05, ** P<0.005. (E) The percentage of polioviruses surviving amiloride treatment relative to untreated control populations was determined by plaque assay. The mean values ± S.E.M are shown, N = 3. ** P<0.005.