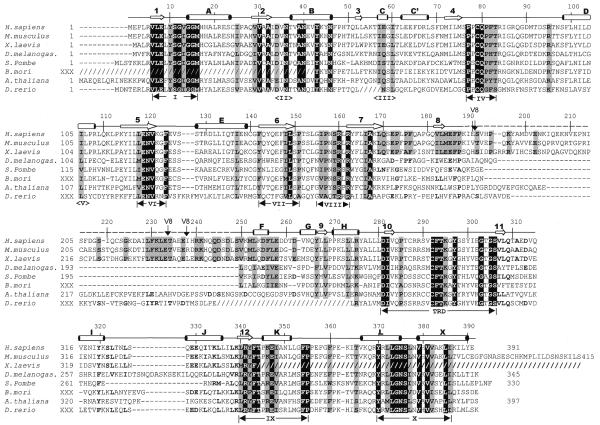
Figure 1.
Alignment of Dnmt2 homologs. The slashed lines indicate sequences missing from the available ESTs and dashed lines indicate gaps introduced to optimize alignments. The human DNMT2 residue numbering is shown above the sequence alignment. Amino acids highlighted are either invariant (white against black) among the proteins or similar (shaded), as defined by the following groupings: V, L, I and M; F, Y and W; K and R; E and D; Q and N; S and T; A, G and P. The secondary structural elements of DNMT2 (helices A–X and strands 1–11) and conserved sequence motifs are labeled. Complete Dnmt2 sequences are available in GenBank for human (AAC39764), mouse (AAC53529) and S.pombe (CAA57824). The two Dnmt2 sequences of D.melanogaster currently deposited in GenBank, AAF03835 and AAF53163, contain a single exon and lack the consensus motif I. We used the sequence reported by Tweedie et al. (24), which includes an additional upstream exon and contains motif I. The A.thaliana sequence was assembled from genomic sequence AC006601 between nucleotides 29989 and 27859. Two of the eight splicing junctions are confirmed by EST sequences. The X.laevis Dnmt2 sequence was assembled from two overlapping ESTs (AW639976 and AW636415) and lacks the C-terminus of the protein. The D.rerio sequence was assembled by jointing four ESTs (AI331437, AI618465, AI331180 and AI331323). The B.mori sequence was assembled from three ESTs (AU005443, AV399933 and AV400288). Shown in dashed overline and labeled V8 are the sequences (from 191 to 237) deleted to make a version of DNMT2 that could be crystallized. Note that these sequences are normally absent from the D.melanogaster, S.pombe and B.mori Dnmt2 homologs.