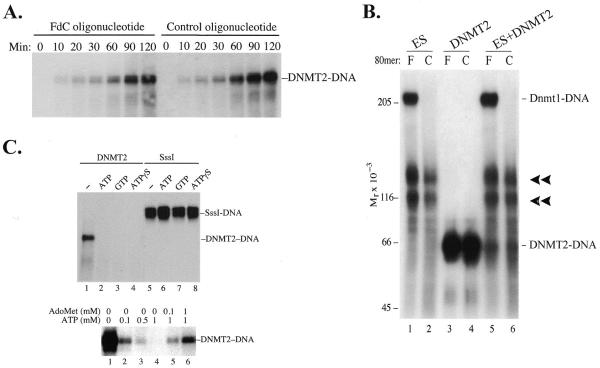
Figure 5.
(A) Formation of denaturant-resistant DNMT2–DNA complexes. DNMT2 (4 µg/ml) was incubated at 37°C with oligonucleotides (1 µg/ml) in 20 mM Tris–HCl pH 8.0, 1 mM EDTA, 25 mM NaCl, 0.2 mM PMSF, 0.5 mM DTT and 40 µM AdoMet. Reactions were terminated at the indicated times by addition of SDS to 2% and glycerol to 12% and heating to 65°C for 10 min, subjected to 6% SDS–PAGE, transferred to nitrocellulose and autoradiographed. Oligonucleotides were of sequence 5′-CCTTTACAAATTTCCAATGCNNNNFGNNNNNNNNFNNNNNNNNNFGNNNNCCTGAAAAAAGACTAATTAAATTCATGGTA-3′, where N is a random base. The synthesis, radioactive labeling and purification of FdC oligonucleotides have been described (16). The control oligonucleotide duplexes were not methylated, while FdC duplexes were hemimethylated (16). Methylation status did not affect formation of complexes (data not shown). Lanes headed F contain FdC oligonucleotides and lanes headed C contain control oligonucleotides with C replacing FdC. (B) Effect of ES cell lysates on formation of DNMT2–DNA complexes. Aliquots of 70 µg ES cell lysate were incubated with 0.1 µg internally labeled FdC or control oligonucleotide in 100 µl of 20 mM Tris–HCl pH 7.4, 10% glycerol, 1 mM EDTA, 0.5 mM DTT, 0.2 mM PMSF and 40 µM AdoMet. Where indicated, purified DNMT2 (0.4 µg) was added at the beginning of the reaction. Reactions were terminated after 3 h at 37°C by addition of SDS to 2% and heating to 65°C for 10 min, subjected to 6% SDS–PAGE, transferred to nitrocellulose and autoradiographed. (C) Inhibition of DNMT2–DNA complex formation by nucleoside triphosphates. Incubation of DNMT2 with FdC oligonucleotide and 1 mM ATP, GTP or ATPγS prevented complex formation but had no effect on the reaction of M.SssI with the FdC oligonucleotide. The inhibitory effect could be largely overcome by addition of equimolar amounts of AdoMet (lanes 5 and 6). In all cases electrophoresis was halted just after unbound oligonucleotides migrated off the lower end of the gel.