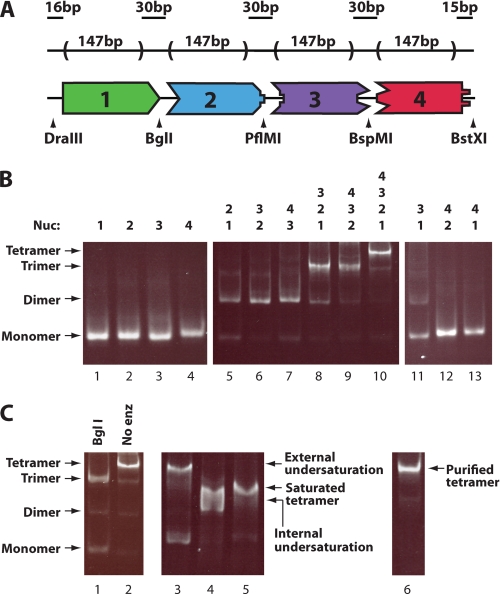
FIGURE 1.
Generation of tetranucleosomal arrays containing specific histone octamers at defined positions. A, shown is the schematic of the tetranucleosomal array DNA template containing four 601-based nucleosome positioning sequences joined head-to-tail by unique nonpalindromic endonuclease cut sites. Arrowheads indicate the location of designated nonpalindromic cut sites. B, shown is characterization and ligation of the four unique mononucleosomes containing wild type octamer. Fragments utilized are shown above the gel. Unligated mononucleosomes (left panel) and mononucleosomes ligated in combinations expected to result in ligated product (center panel) or those that were not (right panel) are shown. Samples were run on a native 4% polyacrylamide gel and stained with ethidium bromide. C, determination of tetranucleosomal array ligation order, saturation, and purity by native gel analysis is shown. Ligated tetranucleosomal array digested with BglI shows expected monomer and trimer products (lane 1). Ligated tetranucleosomal arrays were made harboring an external position of undersaturation (ligation of mononucleosome one, two, and three, with DNA fragment four), an internal position of undersaturation (ligation of mononucleosomes one, two, and four, with DNA fragment three), or with all positions fully saturated (ligation of mononucleosomes 1–4) and are shown in lanes 3–5, respectively. A purified, fully saturated, tetranucleosomal array is shown in the right panel. Nucleosomal array markers shown on the left apply to all six gel lanes.