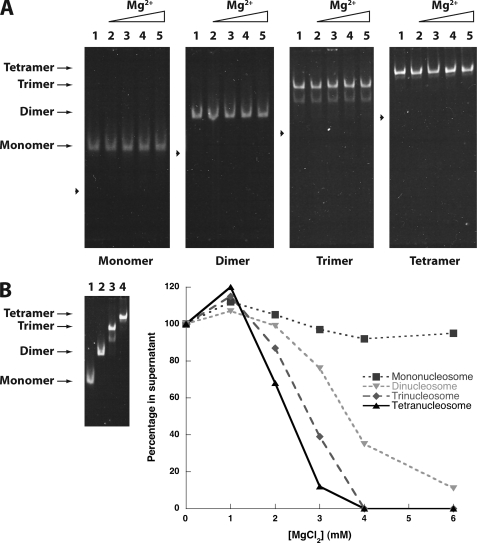
FIGURE 6.
Neighboring nucleosomes enhance intermolecular compaction. A, shown is the presence of detergent enhances nucleosome stability. Mono-, di-, tri-, and tetranucleosomal arrays were incubated in the presence of 0.1% Triton X-100 and increasing concentrations of MgCl2, then run on a native 4% polyacrylamide gel and stained with ethidium bromide. Lanes 0, 0.5, 1.0, 1.5, and 2.0 were incubated in the corresponding mm concentrations of MgCl2. Arrowheads indicate the position of free DNA for each nucleosomal length tested. B, increasing nucleosomal array length enhances intermolecular compaction. Left, mono-, di-, tri-, and tetranucleosomal arrays were incubated in the presence of 0.1% Triton X-100 and 6.0 mm MgCl2, then run on a native 4% polyacrylamide gel and stained with ethidium bromide. Lanes 1–4 show mono-, di-, tri-, and tetranucleosomal arrays, respectively. Right, a representative self-association plot of mono, di-, tri-, and tetranucleosomal arrays is shown. Nucleosomes were incubated with 0.1% Triton X-100 and increasing concentrations of MgCl2. The fraction of nucleosome remaining in the supernatant is plotted as a function of the array concentration.