Table 5.
Estimates of sire polygenic and residual variances using the Bayesian genome-wise model, as a proportion of estimates obtained by ASReml without SNPs
| Genome-wise1 | ASReml2 | ||||
|---|---|---|---|---|---|
| with SNP effect | without SNP effect | ||||
| Trait3 | Polygenic | Residual | Polygenic | Residual | h2 |
| EAH | 0.29 | 0.91 | 0.89 | 1.13 | 0.05 |
| E3 | 0.51 | 1.05 | 1.01 | 0.90 | 0.13 |
| EEW | 0.60 | 1.33 | 0.97 | 1.13 | 0.44 |
| EPD | 0.52 | 1.00 | 1.06 | 0.97 | 0.05 |
| EPS | 0.65 | 1.14 | 0.97 | 1.04 | 0.08 |
| SM | 0.54 | 1.19 | 0.99 | 1.09 | 0.18 |
| EYW | 0.44 | 1.07 | 1.01 | 0.96 | 0.16 |
| LAH | 0.56 | 1.03 | 1.01 | 0.99 | 0.56 |
| LEW | 0.46 | 0.97 | 1.00 | 1.01 | 0.25 |
| LPD | 0.42 | 0.94 | 0.96 | 1.05 | 0.21 |
| LPS | 0.48 | 0.94 | 0.98 | 1.01 | 0.19 |
| LYW | 0.50 | 1.36 | 1.18 | 1.14 | 0.84 |
1 Polygenic and residual variances for each trait were estimated using genome-wise model (MCMC method) with or without fitting SNP effect in the model and presented as proportional to the polygenic and residual variances estimated using ASReml.
2 Polygenic and residual variances for each trait were estimated using ASReml and used for estimating heritability, 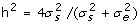 .
.
3 Trait abbreviations: Early Albumen Height (EAH), First 3 Egg Weight (E3), Early Egg Weight (EEW), Early Production (EPD), Early Shell Quality (EPS), Sexual Maturity (SM), Early Yolk Weight (EYW), Late Albumen Height (LAH), Late Egg Weight (LEW), Late Production (LPD), Late Shell Quality (LPS) and Late Yolk Weight (LYW).
