Abstract
Background
H3K9 trimethylation (H3K9me3) and binding of PcG repressor complex-1 (PRC1) may play crucial roles in the epigenetic silencing of the p16 gene. However, the mechanism of the initiation of this trimethylation is unknown.
Methodology/Principal Findings
In the present study, we found that upregulating the expression of PRC1 component Cbx7 in gastric cancer cell lines MGC803 and BGC823 led to significantly suppress the expression of genes within the p16-Arf-p15 locus. H3K9me3 formation was observed at the p16 promoter and Regulatory Domain (RD). CBX7 and SUV39H2 binding to these regions were also detectable in the CBX7-stably upregulated cells. CBX7-SUV39H2 complexes were observed within nucleus in bimolecular fluorescence complementation assay (BiFC). Mutations of the chromodomain or deletion of Pc-box abolished the CBX7-binding and H3K9me3 formation, and thus partially repressed the function of CBX7. SiRNA-knockdown of Suv39h2 blocked the repressive effect of CBX7 on p16 transcription. Moreover, we found that expression of CBX7 in gastric carcinoma tissues with p16 methylation was significantly lower than that in their corresponding normal tissues, which showed a negative correlation with transcription of p16 in gastric mucosa.
Conclusion/Significance
These results demonstrated for the first time, to our knowledge, that CBX7 could initiate H3K9me3 formation at the p16 promoter.
Introduction
Epigenetic silencing is the main way for inactivation of tumor suppressor genes such as p16 (Ink4a, CDKN2A) in numerous human cancers [1]. Both DNA methylation and histone modifications could suppress transcription of these genes. Epigenetic inactivation of p16 is an intensively studied event, which plays a role in carcinogenesis [2]–[4]. Unfortunately, the mechanisms of epigenetic inactivation of such genes including p16 are still largely unknown.
Expression of p16 is regulated by a number of transcription factors, including activator Ras, Myc, TGFβ, ETS2, and silencer pRB, Polycomb group (PcG) proteins such as EZH2, BMI1, CBX2, CBX7, CBX8, etc [5]. Different Polycomb group complexes regulate common target genes [6]. For instance, PcG proteins suppress expression of Homeobox genes and the p16-Arf-p15 locus mainly through the Polycomb repressive complex-1 and -2 (PRC1 and PRC2), which control cell fate and cancer development [6]–[9]. It is well known that binding of PRC2 with the p16 promoter can initiate reversible repression of its transcription through EZH2-mediated trimethylation of histone H3 at lysine 27 (H3K27me3) [10]–[12]. Unlike the reversible H3K27me3, trimethylation of histone H3 at lysine 9 (H3K9me3) is an epigenetic landmark of the eukaryotic genome [13]–[15]. Some PRC1 members such as CBX7 bind to H3K9me3 within both heterochromatin and silenced euchromatic loci in vitro, which might play a role in the epigenetic maintenance of transcriptional silence of target genes [16]. SUV39H1, SUV39H2, and G9a catalyze trimethylation of H3K9 in mammalian cells [17]–[19]. It is reported that CBX7 suppresses transcription of p16 in human fibroblast cells [20], but how CBX7 represses the transcription is not clear. In the present study, we first report, to our knowledge, that the PRC1 member CBX7 could initiate the formation of H3K9me3 at the p16 promoter and thus promote the repression of P16 expression through a SUV39H2/G9a-dependent pathway.
Results
Upregulation of CBX7 suppresses transcription of p16 through initiation of H3K9 trimethylation
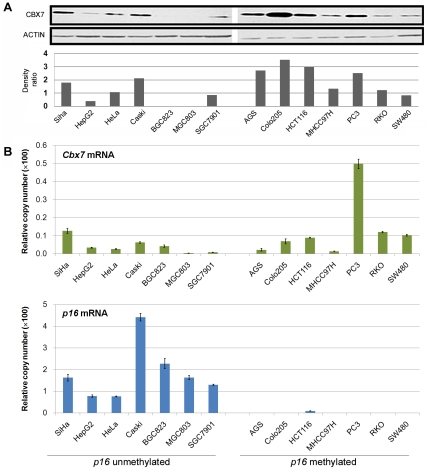
To select suitable cell lines to investigate the role of CBX7 in epigenetic inactivation of p16, we analyzed the methylation status of p16 and the mRNA levels of Cbx7 and p16 in 14 human cell lines (Figure 1 and Fig. S1). p16 Methylation was observed in 6 cell lines including PC3 and HCT116, but not in the gastric cancer cell lines MGC803 and BGC823. Thus, these cancer cell lines MGC803, BGC823 (p16-active and low level of CBX7 expression), and PC3 (p16-methylated and high level of CBX7 expression) were used as the representative cell lines in the subsequent experiments.
Figure 1. CBX7 expression and transcription of Cbx7 and p16 in human cancer cell lines.
(A), analysis of CBX7 expression by Western blot. (B) analysis of of Cbx7 (up) and p16 (down) expression by quantitative RT-PCR. The methylation status of p16 CpG island is marked according to the results displayed on Figure S1.
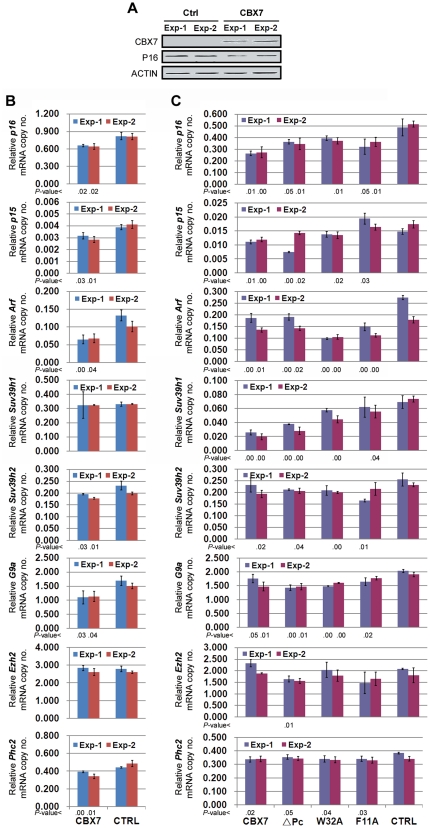
To set up models for silencing p16 transcription by CBX7, we transiently transfected the wildtype Cbx7 expression vector into MGC803 and BGC823 cells. Expression of p16 decreased in both cell lines at 72 hours after transfection (Figure 2). Expression of Arf and p15 located within the same locus also decreased (Figure 2B and 2C). These results suggest that CBX7 can repress the expression of the p16-Arf-p15 locus in these cell lines. Moreover, weak downregulation of histone methyltransferase (HMTase) gene G9a was observed in both cell lines 72 hours after CBX7 transfection. Although transcription of Suv39h1 was drastically reduced in MGC803 cells, consistent downregulation was not observed in BGC823 cells. The levels of Suv39h2 and Ezh2 mRNA were not affected in both cell lines. Transcription of Phc2 was downregulated, although to a weak extent (Figure 2B and 2C).
Figure 2. Effects of CBX7 on expression of the p16-Arf-p15 locus, HMTases, and genes at other loci.
Results of two independent experiments were displayed. (A), analysis of expression of Cbx7 in MGC803 cells 72 hours after transfection by Western blot assays. (B), analysis of transcription level of genes located within the p16-Arf-p15 locus, two control genes (Ezh2 and Phc2) located at other loci and three HMTases, in BGC823 cells 72 hours after transfection by quantitative RT-PCR. (C), analysis of transcription level of genes located within the p16-Arf-p15 locus, two control genes (Ezh2 and Phc2) located at other loci and three HMTases, in MGC803 cells 72 hours after transfection by quantitative RT-PCR. P-value less than 0.05 for CBX7/mutants vs. CTRL was listed below each column, respectively. Each column represents the average value of triplicate. The STDEV value was on the top of column. CTRL, cells transfected with the pcDNA3.1(+)/myc-His A control vector; CBX7, cells transfected with the vector containing the full coding region of wildtype Cbx7. ΔPc, Pc-box deleted CBX7; F11A and W32A, CBX7 containing F11A and W32A mutation within chromodomain, respectively.
It is reported that the Pc-box within CBX7 is essential for its binding to Ring1 and this impairs its ability to repress p16 transcription [20], [21]. Mutations in critical residues of the chromodomain (F11A and W32A, Figure S2) or deletion of the Pc-box (ΔPc) inhibited the ability of CBX7 to extend the life span of cells [20]. In the present study, we also found that the deletion of Pc-box or chromodomain mutations only partially abolished the repressive effect of CBX7 on transcription of p16 in the transiently transfected cell lines. Moreover, the chromodomain mutations abolished the downregulation of Suv39h1 in MGC803 cells significantly. But deletion of the Pc-box had no effect on downregulation of Suv39h1 and p15. Downregulation of Arf was affected by the chromodomain mutation F11A (Figure 2C).
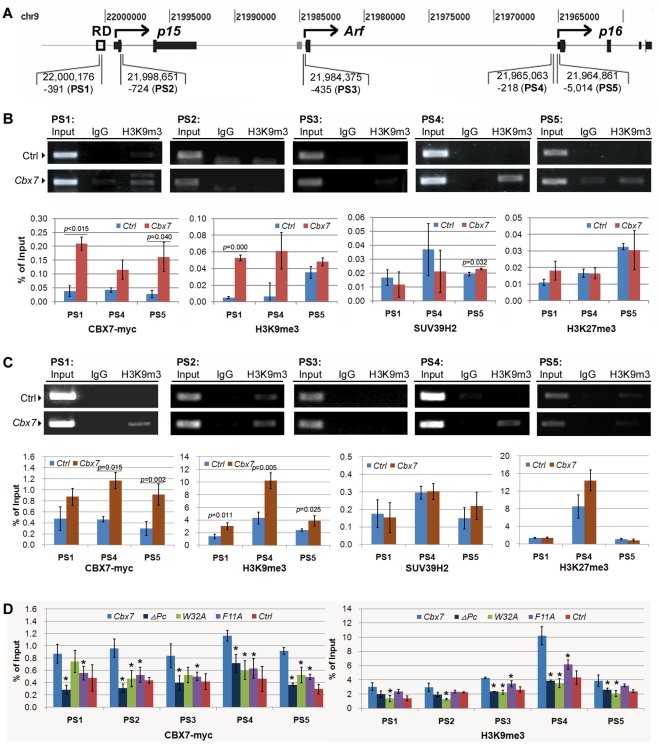
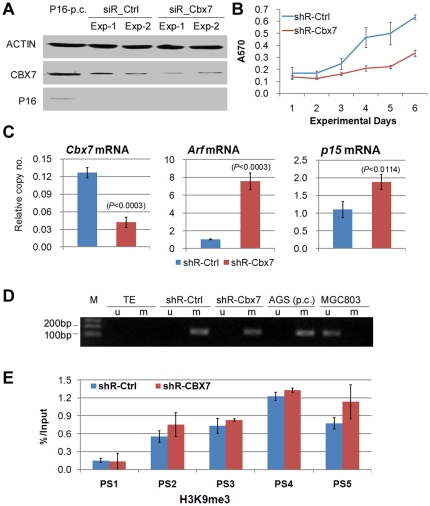
H3K9me2, H3K9me3 and H3K27me3 are well-known epigenetic markers for gene repression. The Regulatory Domain (RD) is reported previously to play an important role in the regulation of p16-Arf-p15 locus [22]. To address whether histone modifications are involved in the repression induced by CBX7, we analyzed H3K9me3 at the p16-Arf-p15 locus and RD using chromatin immunoprecipitation assay (ChIP). H3K9me3 was seen within the RD and the p16 promoter and 5′UTR regions in the Cbx7 transiently transfected MGC803 and BGC823 cells (Figure 3B and 3C: PS1, PS4, and PS5). Formation of H3K27me3 was not increased within these tested loci (Figure 3C). H3K9me2 was not detectable either (data not shown). Binding of CBX7 mutant proteins to five tested DNA fragments within the p16-Arf-p15 locus was not detectable and formation of H3K9me3 was not induced either (Figure 3D).
Figure 3. H3K9me3 formation, CBX7 and SUV39H2 binding within p16 and Regulatory Domain regions in ChIP assays.
(A), locations of the p15-Arf-p16 locus at chromosome 9 and the corresponding fragments amplified with primers (PS1∼5) used in ChIP assays (based on web http://genome.ucsc.edu, Mar. 2006, assembly). (B) and (C), analysis of H3K9me3 formation by ChIP assay in cell line MGC803 and BGC823 transiently transfected with Cbx7 expression vector, respectively. H3K9me3 formation, CBX7- and SUV39H2-binding were also analyzed by quantitative ChIP assays in these cell lines. (D), results of CBX7-binding and H3K9me3 formation by ChIP assays in BGC823 cells 72 hours after transfected with the wildtype and mutant Cbx7-vectors. *, versus the wildtype CBX7, P<0.05; Ctrl, cells transfected with the control vector pcDNA3.1(+)/myc-His A; Cbx7, cells treated with the vector encoding full length of coding region of the wildtype CBX7; ΔPc, Pc-box deleted CBX7; F11A and W32A, CBX7 containing F11A and W32A mutation within chromodomain, respectively; % of Input, the percentage of the average relative copy number of the tested triplicates to that of Input control.
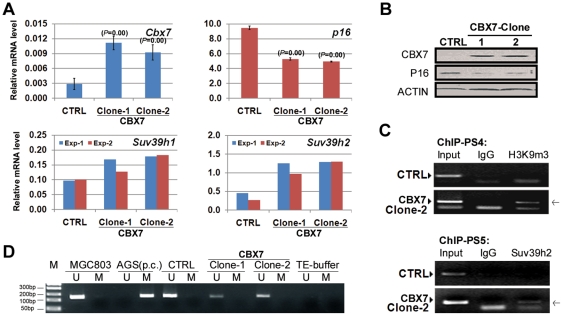
In the CBX7-stably transfected MGC803 subclone(s), in which downregulation of p16 expression is maintained, expression of Suv39h1and Suv39h2 increased significantly (Clone-1 and Clone-2; Figure 4A and 4B). The H3K9me3 formation was shown within the tested p16 promoter (Figure 4C: PS4), though p16 methylation was not observed in these subclones even at passage 80 by methylation-specific PCR (MSP) (Figure 4D: Clone-1 and Clone-2).
Figure 4. HMTase expression, H3K9me3 formation, SUV39H2 binding, DNA methylation within p16 CpG island.
(A), analysis of expression of Suv39h1 and Suv39h2, p16, and Cbx7 in two CBX7-stably transfected MGC803 subclones by quantitative RT-PCR. (B), analysis of CBX7 and P16 expression in the CBX7-stably transfected subclones by Western blot. (C), ChIP analysis of H3K9me3 formation within the p16 promoter and recruitment of SUV39H2 to the 5′UTR of p16 gene in Clone-2. The target PCR product is marked by an arrow. (D), analysis of p16 methylation by methylation-specific PCR (MSP). Genomic DNA of AGS cell line was used as positive control (p.c.) for p16 methylation in the MSP assay. CTRL: MGC803 cell stably transfected with pcDNA3.1(+)/myc-His A control vector; Clone-1/-2: the two MGC803 subclones stably transfected with vector containing the full coding region of wildtype Cbx7; Exp-1/-2: two independent experiments.
In addition, transient knockdown of CBX7 by siRNA did not reactivate P16 expression in p16-methylated PC3 cells (Figure 5A). Stably knockdown of CBX7 by shRNA inhibited the growth of PC3 cells (Figure 5B), but induced neither p16 transcription (Figure 5C) nor demethylation of p16 CpG island (Figure 5D). Significant change of H3K9me3 level at the five tested fragments was not observed (Figure 5E). However, significant upregulation of the transcription of Arf and p15 was observed in PC3 cells (Figure 5C).
Figure 5. Effect of downregulation of Cbx7 on expression of p16/Arf/p15, cell growth and p16 methylation.
(A), analysis of CBX7 expression in PC3 cells by Western blot assay 72 hours after CBX7 knockdown. Protein extracted from 293T cells was used as P16 positive control (P16-p.c.). (B), the growth curves of PC3 cells stably transfected with the scramble control shRNA vector (shR-Ctrl) or shRNA-Cbx7 vector (shR-Cbx7) in MTT assay. (C), analysis of transcription of Cbx7, Arf, and p15 by quantitative RT-PCR. The statistical significance of the differences of mRNA relative copy number between cells stably transfected with shR-Ctrl and shR-Cbx7 were labeled above the top of one of the columns. (D), analysis of p16 methylation by MSP in the Cbx7 stably knockdown PC3 cells. Genomic DNA from AGS and MGC803 cell lines was used as positive control of methylated and unmethylated p16 respectively. (E), comparison of H3K9me3 level by ChIP assays within various locations as illustrated in Figure 3A.
SUV39H2 and G9a contribute to H3K9 trimethylation within the p16 5′UTR induced by CBX7
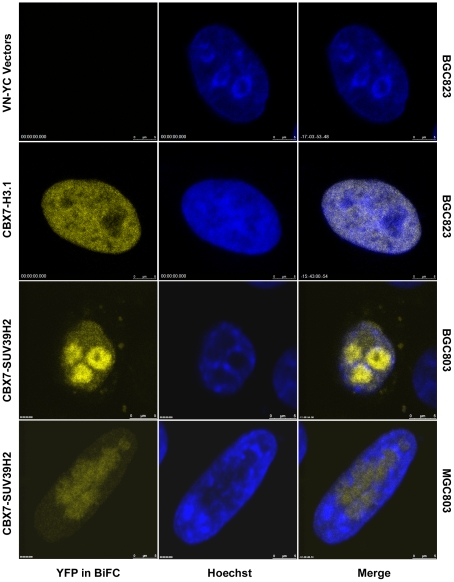
G9a, SUV39H1 and SUV39H2 are three histone methyltransferases (HMTases) involved in trimethylation of H3K9. Hence, we studied which HMTase was the enzyme responsible for the trimethylation of H3K9 after CBX7 upregulation. We initially analyzed the binding of SUV39H2 with p16-Arf-p15 locus by ChIP assay, and found that SUV39H2 was weakly recruited to the p16 5′UTR in cell lines transiently transfected with CBX7 (Figure 3B and 3C), but strongly in the tested CBX7-stably upregulated subclone-2 (Figure 4C). Thus, bimolecular fluorescence complementation assay (BiFC) was further used to detect the possible interaction between CBX7 and SUV39H2. Amino acids 173–238 of YFP (YC) were fused to the C-termini of SUV39H2 to construct the YC-Suv39h2 vector. The VN-Cbx7 vector, containing the N-terminal 172 amino acids of Venus (VN) fused to the N-termini of CBX7 protein, was also used in the BiFC assay, because it remains an active CD domain with high affinity to Histone 3.2 and 3.1 [23]. Images of the BiFC complex of CBX7-SUV39H2 (yellow) and Hoechst (blue) fluorescence were obtained in BGC823 (Figure 6) and MGC803 cells 36 hours upon transfection (Figure S3). Interaction between CBX7 and SUV39H2 was mainly observed within the nucleus, especially the Hoechst-negative staining areas in both cell lines (Figure 6 and Figure S3). Such interaction could not be observed in the tested cells transfected with blank VN- and YC-vector controls. Interaction between CBX7 and Histone 3.1 positive control was mainly observed within the Hoechst-positive staining area (Figure 6). At the same time, we cloned Cbx7 into the pEFGP-C1 vector and transfected it to MGC803 cells and found that CBX7 was distributed in the entire nucleus with higher concentration at the Hoechst-negative staining areas (Figure S3), which was consistent with the BiFC results. Although CBX7-Histone 3.1 complexes were observed in Co-IP assay, CBX7-SUV39H2 complexes were not detected (Figure S4).
Figure 6. Distribution of CBX7-SUV39H2 complexes in cancer cell line BGC823 in bimolecular florescence complementation (BiFC) assay.
36 hours after transfection, images of the BiFC complex of CBX7-SUV39H2 (yellow) and Hoechst (blue) fluorescence were obtained and merged. The blank VN- and YC-vectors were used as negative controls. VN-CBX7 and YC-Histone 3.1 (H3.1) vectors were used as positive controls.
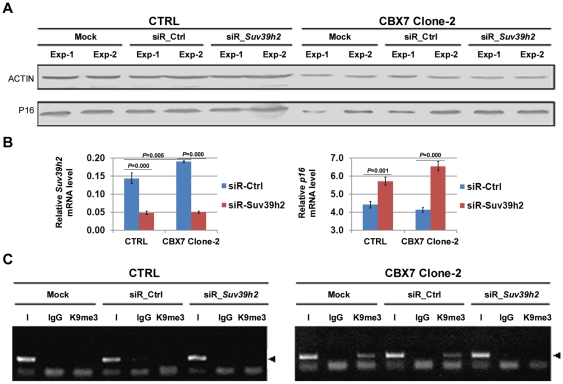
Moreover, a rescue assay was carried out to evaluate the role of SUV39H2 in the formation of H3K9me3 within the p16 promoter induced by CBX7. We found that knockdown of Suv39h2 expression by siRNA abolished the formation of H3K9me3, and therefore rescued p16 expression in the tested subclone (CBX7 Clone-2; Figure 7A∼C). Moreover, decrease of H3K9me3 was also observed after knockdown of G9a by siRNA, but it could not be detected in the tested cells after treatment with siRNA-Suv39h1, although p16 expression was increased after these siRNA treatments (Figure S5). These results together indicate that CBX7 may repress p16 expression by recruiting SUV39H2 to the p16 5′UTR to induce the formation of H3K9me3. In addition, in the p16-methylated PC3 cells, after stably knockdown of CBX7, p16 transcription was still not detectable upon further knockdown of Suv39h2 and/or Suv39h1 expression by siRNAs. Again, knockdown of Suv39h1 led to upregulation of Suv39h2, whereas knockdown of Suv39h2 did not lead to upregulation of Suv39h1 in the PC3 cells (Figure S6).
Figure 7. Effect of siR-Suv39h2 on p16 expression, H3K9me3, and binding of SUV39H2 within the p16 5′UTR.
The CBX7-stably transfected MGC803 subclone CBX7 Clone-2 was analyzed 72 hours after knockdown of Suv39h2 by siRNA. (A), analysis of P16 expression by Western blot. (B), analysis of transcription of Suv39h2 by quantitative RT-PCR. (C), analysis of H3K9 trimethylation (K9me3) within the p16 promoter by ChIP assays. siR_Ctrl, scramble siRNA control; siR_Suv39h2, siRNA targeted to Suv39h2.
Expression alterations of CBX7 and other PRC1 components in human gastric carcinomas
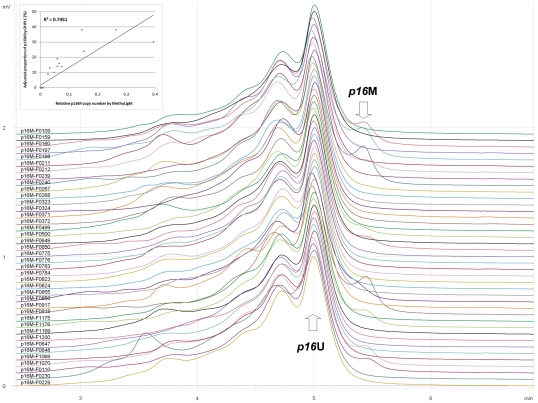
To validate whether epigenetic silence of p16 expression correlates negatively with transcription of Cbx7 and its related components in vivo, we first analyzed p16 methylation status quantitatively by DHPLC and MethyLight in human primary gastric carcinoma samples (GC), the corresponding cutting-edge normal tissues of GC (GCN), and human normal gastric mucosa biopsies from non-cancer patients (Normal). Peak for methylated-p16 (p16M) was observed in eleven gastric carcinoma samples: F0110 (14%), F0160 (24%), F0198 (30%), F0212 (10%), F0240 (38%), F0500 (9%), F0650 (13%), F0856 (14%), F0918 (38%), F1070 (16%), and F1176 (19%) (Figure 8). The results were confirmed by MethyLight (Figure 8, inserted chart) and bisulfite clone sequencing (data not shown).
Figure 8. DHPLC chromatogram of methylation status of p16 CpG island in human primary gastric carcinoma.
20 pairs of gastric carcinomas (with even ID) and their corresponding normal samples (with odd ID) were analyzed by DHPLC. The inserted chart displayed that the result of DHPLC detection correlated with that of quantitative MethyLight analysis well (R2 = 0.745; P = 0.000).
We also analyzed the transcription levels of p16, Cbx7, and other 6 PcG genes in these samples by quantitative RT-PCR (Table 1). Expression of p16 in GCs with p16 methylation was lower than that without p16 methylation, but not significant (Table 1, P = 0.067; the lower ΔCt value, the higher mRNA amount). Transcription of Cbx7 in GCs was lower than that in GCNs, especially in GCs with p16 methylation (P = 0.009). Transcription of p16 in GCNs (and GCs) was significantly higher than in Normal samples (P = 0.003) whereas transcription of Cbx7 in GCNs (and GCs) was significantly lower than in Normal samples (P = 0.039), which suggest an inverse relationship between transcription of p16 and Cbx7 among gastric samples without p16 methylation.
Table 1. Comparison of transcription levels (ΔCt in qRT-PCR assay, a high ΔCt value means low transcription) of p16 and seven PcG genes in different kinds of human gastric mucosa samples with and without p16 methylation.
| PcGs | p16M a | ΔCt in qRT-PCR (Mean±SD) | P-value, vs. GCN | |||
| GC (n = 20) b/ c | GCN (n = 20) | Normal (n = 18) | GC d | Normal | ||
| p16 | Positive | 12.62±2.59 b/ e | 11.34±1.39 f | 0.217 | ||
| Negative | 10.42±2.41 c | 11.80±2.03 | 14.63±1.48 g | 0.243 | 0.003 | |
| (Total) | 11.63±2.41 | 11.55±2.03 | 0.919 | |||
| Cbx7 | Positive | 8.94±1.08 | 7.42±1.37 | 0.009 | ||
| Negative | 7.77±2.23 | 6.94±1.42 | 5.54±1.77 | 0.364 | 0.039 | |
| (Total) | 8.41±1.75 | 7.2±1.37 | 0.019 | |||
| Ring1 | Positive | 6.81±0.82 | 7.29±1.65 | 0.359 | ||
| Negative | 6.67±1.34 | 6.57±1.37 | 6.62±1.38 | 0.886 | 0.927 | |
| (Total) | 6.74±1.06 | 6.97±1.53 | 0.595 | |||
| Bmi1 | Positive | 9.21±0.86 | 9.56±1.29 | 0.459 | ||
| Negative | 8.23±1.66 | 8.58±0.80 | 8.46±2.24 | 0.584 | 0.764 | |
| (Total) | 8.77±1.34 | 9.12±1.18 | 0.389 | |||
| Mel18 | Positive | 4.81±0.88 | 5.82±1.35 | 0.051 | ||
| Negative | 4.86±1.10 | 4.97±1.46 | 5.80±1.63 | 0.871 | 0.198 | |
| (Total) | 4.83±0.96 | 5.44±1.43 | 0.128 | |||
| Phc2 | Positive | 9.21±0.90 | 9.25±1.33 | 0.994 | ||
| Negative | 8.25±1.77 | 8.26±1.16 | 9.33±2.3 | 0.991 | 0.213 | |
| (Total) | 8.78±1.42 | 8.8±1.32 | 0.959 | |||
| Cbx8 | Positive | 9.17±0.68 | 9.38±0.47 | 0.420 | ||
| Negative | 8.50±0.93 | 9.09±0.72 | 8.04±1.34 | 0.149 | 0.014 | |
| (Total) | 8.87±0.85 | 9.25±0.6 | 0.110 | |||
| Ezh2 | Positive | 5.38±1.19 | 8.90±1.53 | 0.000 | ||
| Negative | 6.53±1.73 | 6.72±1.66 | 6.61±2.27 | 0.816 | 0.885 | |
| (Total) | 5.90±1.53 | 7.92±1.90 | 0.001 |
GC: human primary gastric carcinoma sample; GCN: the corresponding cutting- edge tissues of GC; Normal: human normal gastric mucosa biopsies from non-cancer patients;
methylation of p16 CpG island (p16M) in gastric mucosa tissues from patients with or without cancer;
GC samples from 11 patients are p16M-positive;
GC samples from 9 patients are p16M-negative;
paired t-test;
p16M-positive GCs vs. p16M-negative GCs, P = 0.067;
p16M was detectable in GCs, but not in the corresponding GCN by DHPLC;
p16 mRNA was detectable in 12 of 18 Normal gastric biopsies; all of 18 Normal samples are p16M-negative.
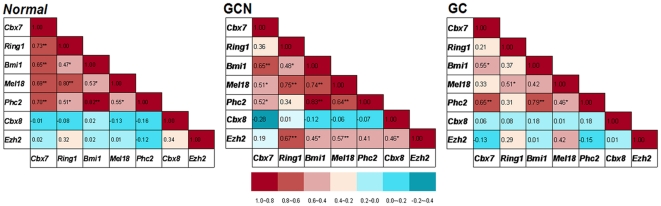
Interestingly, in Normal tissues from 18 non-cancer patients, we found that transcription of Cbx7 was positively correlated with those of Ring1, Bmi1, Mel18, and Phc2, but not with those of Cbx8 and Ezh2 (Figure 9, left chart; Ps<0.01). However, in GCN and GC samples from 20 cancer patients, such correlation was progressively disturbed, especially between Cbx7 and Ring1/Mel18 (Figure 9, middle and right chart). Moreover, in contrast to Cbx7 expression, the average expression levels of Ezh2 in Normal, GCs and GCNs were similar in the samples without p16 methylation, but transcription of Ezh2 in GCs with p16 methylation was significantly higher than that in GCNs (P<0.000). These results suggest that expression patterns of PcGs, especially CBX7, are changed during development of gastric carcinoma and may affect p16 expression in the human stomach in vivo.
Figure 9. Correlation coefficients between mRNA levels of PcG genes in various kinds of gastric mucosa samples.
The relative copy numbers (RCN) was calculated based on the average Ct number of target gene and the GAPDH reference [2-(Cttarget_gene-CtGAPDH)]. The correlation coefficients was calculated based on each gene's RCN for each sample within different groups, including normal gastric mucosa biopsies (Normal, N = 18), gastric carcinomas (GC, N = 20) and their corresponding normal samples (GCN). The significance of correlation coefficients was calculated using the statistic software SPSS16.0 and marked with */** (P<0.05/0.01).
Discussion
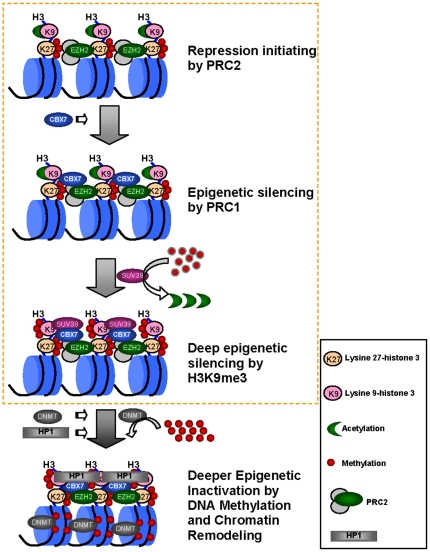
Epigenetic inactivation of p16 is a well-studied event during carcinogenesis. The detailed mechanism of p16 inactivation by DNA methylation and histone modification is still unclear. It is well known that H3K9me3 and PRC1 formation in the silenced euchromatic loci, such as the methylated p16-Arf locus, are epigenetic inactivation landmarks (Figure 10). Heterochromatin binding protein HP1 can bind with H3K9me3 [13]. Interactions between the PRC1 member CBX7 and H3K9me3 are also observed in in vitro peptide pull-down assays [16]. However, it is largely unclear how the locus specific H3K9 trimethylation is initiated during its epigenetic silence processes. In the present study, we first observed, to our knowledge, that the possible H3K9me3 binding protein CBX7 itself could initiate the specific formation of H3K9me3 via HMTase SUV39H2-dependent pathways in the p16 locus within gastric cancer cell lines.
Figure 10. Epigenetic silencing pathway for the p16-Arf-p15 locus.
Gene repression pressure leads to recruitment of EZH2-containing Polycomb repressive complex-2 (PRC2) to the transcription complex, that trimethylates H3K27 and thus initiates the repression. The PRC2-related repression is reversible under condition of transcription pressure. However, in the case of long-term silence, Polycomb repressive complex-1 (PRC1) is recruited to the PRC2-repressed genes and activates H3K9 trimethylation subsequently, which may subsequently induce DNA methylation and chromatin remodeling with assistance of other silencers including DNMTs, HDACs, and heterochromatin binding protein HP1. The content within the yellow dash-square presents the discovery in the present study that connects two epigenetic landmarks the PRC1 binding and H3K9 trimethylation together.
It is reported that CBX7 promotes proliferation of several human cell lines by repression of p16 expression [6], [20], [21]. To understand the possible mechanisms through which CBX7 represses p16 transcription, we established CBX7 overexpression models by transfecting wildtype CBX7 expression vector to MGC803 and BGC823 cells and found both transiently and stably upregulated CBX7 could repress p16 expression in these gastric cancer cell lines, increase levels of H3K9me3 within the p16 promoter and RD regions. But an increase of H3K27me3 and H3K9me2 was not detectable within the same regions. CBX7 binding to these regions suggests that CBX7 may repress p16 expression through initiating or protecting trimethylation of H3K9. Although consistent recruitment of SUV39H2 to the p16 promoter and 5′UTR regions is not observed in two CBX7 transiently upregulated cell lines, it is found that SUV39H2 is not only upregulated, but also recruited to the p16 5′UTR in the CBX7 stably transfected subclones. This indicates that SUV39H2 may be involved in the maintenance of the established H3K9me3 pattern. Like CBX7-GFP, main interaction between CBX7 and SUV39H2 was observed in the Hoechst negative staining area in BiFC assay. Weak interactions of CBX7 with SUV39H2 were also observable in the Hoechst positive staining area. Both are consistent with the distribution of CBX7. These results indicate that CBX7 and SUV39H2 may function together physically within chromatin in the transfected cells. CBX7 could interact with Histone 3.2 or 3.1 directly in BiFC assay [23]. Such interaction between CBX7 and H3.1 in nucleus was also observed in the BiFC assay. Moreover, we found the complex of Myc-CBX7 and YC-H3.1 in Co-IP assay. Unfortunately, we did not find the complex of Myc-CBX7 and YC-SUV39H2 in the Co-IP assay. The lower detection sensitivity of Co-IP compared to that of the BiFC assay and weak interaction between CBX7 and SUV39H2 might account for the result difference. Taken together, it is reasonable to make the assumption that SUV39H2 may responsible for the formation of H3K9me3 at the p16 promoter. It supports our hypothesis that knockdown of SUV39H2 inhibited the formation of H3K9me3 and rescued p16 expression. Another HMTase G9a, but not SUV39H1, might also contribute to the H3K9 trimethylation. We conclude that CBX7 could initiate or maintain H3K9 trimethylation at the p16 locus in a SUV39H2/G9a-dependent pattern. Although we did not observe CBX7-induced H3K9me3 formation within the p15 and Arf promoter, we cannot exclude that CBX7 induces trimethylation of other genes in the SUV39H2-dependent way.
CBX7 protein contains a C-terminal Pc-box and a N-terminal chromodomain. The binding of the Pc-box to Ring1 is necessary for the repression of p16 expression. The chromodomain plays an important role in the binding of CBX7 to H3K9me3 in heterochromatin and silenced euchromatic loci [16], [20], [21]. Mutations in critical residues of chromodomain (F11A and W32A) or deletion of the Pc box (ΔPc) inhibited the ability of CBX7 to extend the life span [20]. In the present study, we found that all of these mutants only partially abrogated the repressive function of CBX7 on p16 expression, because the residual function of these mutants was still significant. Most importantly, we found that CD mutations or deletion of the Pc-box of CBX7 abolished binding of CBX7 to the tested DNA fragments and greatly disrupted the induction of H3K9me3 within the p15-Arf-p16 locus in ChIP assays. These results demonstrate that both functional motifs within CBX7 protein contribute to the repression of the p15-Arf-p16 locus through the H3K9me3 formation.
Mouse CBX7 displays strong affinity for both H3K9me3 and H3K27me3 in vitro and is developmentally regulated in its association with chromatin [16], [23]. In the present study, we did not find an increase of H3K27me3 formation within the p16 promoter. The HMTase EZH2 is responsible for H3K27 trimethylation. Upregulation of Ezh2 was not observed in the Cbx7 upregulated cells either. Whether CBX7 binding to H3K27me3 is a prerequisite for recruitment and/or activation of SUV39H2 to the p16 promoter and subsequent trimethylation of H3K9 is unclear.
Recently, it is reported that enforced upregulation of CBX7 promotes E-cadherin expression in 293T cells through its physical interaction with HDAC2 and inhibited its activity within the E-cadherin promoter [24]. In contrast, CBX7 could bind to DNMT1 and induce methylation of E-cadherin and other tumor suppressor genes, not including p16, in the embryonal carcinoma cell line Tera-2 [25]. G9a-DNMT complexes were also detected in colorectal cancer cell line RKO [25]. In the present study, we did not analyze changes of epigenetic modifications within the E-cadherin promoter because E-cadherin is methylated in both MGC803 and BGC823 cell lines [26]. We analyzed the p16 promoter and did not find methylation of CpG islands of p16 in the CBX7 stably transfected cell lines even at passage 80. Instead, H3K9me3 formation was detected in the p16 5′ UTR, which may account for the downregulation of p16 transcription. This difference may be due to the different cell lines used in each study. It also implies that CBX7 have multiple functions and thus regulate gene expression through different mechanisms. Additionally, we also found that stable knockdown of CBX7 by shRNA did not induce de-repression of p16 expression and demethylation of p16 CpG island in PC3 cell line either even after Suv39h2 and/or Suv39h1 were knockdown by siRNAs. Similar results in RKO cell line were also reported [25]. In consideration of above observation that CBX7 is downregulated in primary gastric carcinomas with p16 methylation and CBX7 knockdown does not induce demethylation of p16 in PC3 cells, it is likely that CBX7 upregulation may be sufficient for induction or maintenance of H3K9me3, but not for the establishment of DNA methylation within the p16 locus. Because of the close relationship between H3K9me3 and initiation of DNA methylation, additional studies will reveal whether CBX7 upregulation is necessary for initiation of DNA methylation of p16.
We found that upregulation of CBX7 decreased transcription of Arf and p15, while downregulation of CBX7 increased transcription of Arf and p15, which is the same as p16 gene. Unlike within p16, H3K9me3 formation within the Arf and p15 loci was not observed in the tested cancer cell lines after transiently transfection of CBX7. Correlation between Cbx7 expression and Arf methylation/transcription was not observed in the present study either (data not shown). These phenomena imply that CBX7 may favor to repress p16 more than it does Arf and p15. It is reported that RD regulates expression of the p15-Arf-p16 locus simultaneously [5], [22] and we observed H3K9me3 formation at RD region after CBX7 transfection. Thus, formation of H3K9me3 within the RD might account for the downregulation of p15 and Arf transcription induced by CBX7.
Polycomb proteins such as EZH2, BMI1, CBX8, and CBX7 may play important roles in suppressing the expression of the p16-Arf locus in vitro [6], [7], [12], [27], [28]. However, which kind of PcG protein may contribute to epigenetic silence of this locus in vivo is not evaluated yet. Others have reported downregulation of Cbx7 in human cancer tissues previously [28]–[31]. The relationship between transcription of Cbx7 and p16 is not evaluated in gastric tissues. To validate the repression effect of CBX7 on transcription of p16 in vivo, the relationships between transcription of p16, Cbx7 and other PRC1 components were analyzed using Normal gastric mucosa biopsies from 18 non-cancer patients and paired GCs and GCNs from 20 cancer patients (Table 1). We found that transcription of Cbx7 was significantly lower in GCs than in GCNs, especially among samples from patients with GCs containing p16 methylation. Furthermore, we observed that Cbx7 expression was negatively correlated with p16 expression: transcription level of Cbx7 was high in Normal tissues with low level of p16 transcription, but it was low in GCNs and GCs with high level of p16 transcription. This provides in vivo evidence that CBX7 may be involved in the repression of p16.
In addition, we also found for the first time that transcription of Cbx7 was significantly correlated with those of other PRC1 components Bmi1, Mel18, Ring1, and Phc2 in Normal gastric tissues, while not with those of Cbx8 and Ezh2. However, such correlations were progressively disturbed in GCNs and GCs.
Downregulation of Cbx7 in GCs and GCNs may account for disruption of these correlations. It was reported that MEL18 could repress BMI1 in cancer cells [32] and knockdown one of PcGs might release other PcGs from its target gene (such as p16) [7]. We proposed that the correlated expression pattern of these PcGs in normal gastric tissues might be necessary for maintenance of their normal functions, and disruption of the pattern might result in abnormal repression of their target genes.
In conclusion, we found that the PRC1 member CBX7 could initiate trimethylation of H3K9 at the p16-Arf locus through recruitment and/or activation of the HMTase SUV39H2 to the target locus. This finding links two repressive epigenetic landmarks H3K9me3 formation and PRC1 binding within the silenced domains in euchromatin together, and builds up a full pathway for epigenetic inactivation of p16 by histone modifications (Figure 10).
Materials and Methods
Cell lines and human gastric samples
Human cell lines BGC823, HepG2, MGC803, RKO, and SGC7901 were cultured in RPMI1640 medium with 10% FBS. Human cell line Caski, Colo205, HCT116, HeLa, MHCC97H, Siha, and SW480 were cultured in DMEM medium with 10% FBS. AGS and PC3 were cultured in F12 medium with 10% FBS. All of these cell lines were cultured at 37°C with 5% CO2. PC3 cell line was purchased from Cell Line Bank, Chinese Academy of Medical Science. RKO cell line was kindly provided by Dr. Guoren Deng at University of California San Francisco. SW480 and HCT116 cell lines, by Dr. Yuanjia Chen at Peking Union Hospital. HepG2 (from Qingyun Zhang), MGC803, BGC823, SGC7901, HeLa, Caski, Siha, and other cell lines (from Dr. Yang Ke) were obtained from laboratories at Beijing Cancer Hospital/Institute. When the density of cells in the dish was closed to 70%, cells were harvested for extraction of proteins, mRNA, and DNA samples as described below. Proliferation of cells was analyzed with MTT assay.
Twenty paired primary gastric carcinoma (GC) and their corresponding cutting-edge normal tissue (GCN) samples from GC patients (11 males and 9 females, 33–74 years old, the average age 59-y) and 18 gastric normal biopsies (Normal) from non-cancer patients (with or without mild/moderate chronic gastritis) were collected and fresh-frozen at −70°C at Beijing Cancer Hospital. All clinical samples and histopathological information for each case were obtained according to approved institutional guidelines.
Ethics Statement
Informed consent was obtained from all subjects and the institutional review committee approved this study.
Plasmid construction and transfection
Coding region of CBX7 (GenBank accession no. XM_066324) was cloned into pcDNA3.1(+)/myc-His A vector (Invitrogen) to get the wildtype CBX7 expression vector. To construct mutant vectors, point mutations of caging aromatic residues (F11A and W32A; Figure S2) within the chromodomain of CBX7 and the Pc-box (231–242 amino acids)-deleted CBX7 (ΔPc) were created using Easy Mutagenesis System (FM101; Transgene). These plasmids were transfected to MGC803 and BGC823 cells using Lipofectamine™ 2000 (11668-027; Invitrogen) according to the manufacturer's instruction, respectively.
ShRNA against Cbx7 was constructed by oligonuleotide targeted to Cbx7 mRNA (110 nt–128 nt: 5′-caaag tacag cacgt ggga-3′) (accession number XM_066324) in pGFPU6/Neo shRNA expression vector including GFP coding sequence (GenePharma Company, Shanghai). The scramble shRNA (5′-gttct ccgaa cgtgt cacgt-3′) was used as negative control. These plasmids were transfected to PC3 cells.
Generation of the stable cell clones
Wildtype CBX7 expression vector and the control vector were transfected to MGC803 cells. After transfection, cells were cultured in selective medium containing G418 (700 µg/ml). Two subclones (Clone-1 and Clone-2) stably expressing CBX7 and one control cell clone were obtained.
PC3 cells with stable knockdown of Cbx7 were obtained by sorting twice with FACS as followed. 24 hours after transfection, PC3 cells were cultured in the selective medium containing 300 µg/ml of G418 for 3 weeks. Then, these cells were sorted by FACS. GFP-positive cells were collected and cultured in the selective medium, and sorted again after 3 weeks' culture.
Protein preparation and Western blot analysis
Total protein was extracted from cultured cells or fresh tissue samples with TRIzol reagent (Invitrogen) according to the manufacturer's instruction. Immunoblotting was performed with the following antibodies: rabbit anti-CBX7 (Ab21873; Abcam), mouse anti-P16 (Ab50282; Abcam), and mouse anti-β-ACTIN (Santa Cruz). Chemiluminescent reagent (Pierce, Thermal) was used to display the protein bands transferred on the PVDF membrane (Millipore).
RNA extraction, RT-PCR, and quantitative RT-PCR (qRT-PCR) assays
Total RNA was isolated using RNeasy mini kit (Cat. #74104, QIAGEN) according to the manufacture's instruction. Total RNA was reverse transcripted into cDNA using ImProm-II TM Reverse Transcription System (A3800; Promega). Quantitative RT-PCR was performed using power SYBR Green PCR Master Mix (P/N 4367659, Applied Biosystems) on an ABI-7500 Real-Time PCR system (Applied Biosystems). Cbx7 mRNA level was quantified with TaqMan kit (Roche). The primer sets for the tested genes were listed in Table 2. The relative gene mRNA level was calculated based on the average Ct number of target gene and the GAPDH reference [2-(Cttarget_gene-CtGAPDH)]. SPSS16.0 software was used to analyze the data.
Table 2. Primer sequences.
| Gene name | Entrez Gene | Assay | Oligo name | Primer Sequence (5′→3′) | PCR Products | Tm (°C) | References |
| Cbx7 | 23492 | RT-PCR | Cbx7-F | aaagtcgagtatctggtgaagtgg | 442bp | 66 | |
| Cbx7-R | gctcccgtcgatggctgtgg | ||||||
| qRT-PCR | Cbx7-qF | cgtcatggcctacgagga | 71bp | 54 | Pallante et al, 2008 | ||
| Cbx7-qR | tgggtttcggacctctctt | ||||||
| Cbx7-qProbe | aggaggag | ||||||
| Cbx8 | 57332 | qRT-PCR | Cbx8-qF | ctcgtgaaatggaagggatggt | 222bp | 53 | |
| Cbx8-qR | gatgcccctggctgagtcact | ||||||
| Bmi-1 | 648 | qRT-PCR | Bmi1-qF | aattagttccagggcttttcaa | 120bp | 60 | |
| Bmi1-qR | cttcatctgcaacctctcctctat | ||||||
| Ezh2 | 2146 | qRT-PCR | Ezh2-qF | cggggatagagaatgtgggttta | 173bp | 60 | |
| Ezh2-qR | aggtgggcggctttctttatcatc | ||||||
| Mel18 | 7703 | RT-PCR | Mel18-qF | tggggatggggacaaagagaaaac | 200bp | 60 | |
| Mel18-qR | tccgccgccaggggtagat | ||||||
| Suv39h1 | 6839 | qRT-PCR | Suv39h1-qF | ggcaacatctcccactttgt | 250bp | 56 | |
| Suv39h1-qR | caatacggacccgcttctta | ||||||
| Suv39h2 | 79723 | qRT-PCR | Suv39h2-qF | cccatctatgaatgcaactcaag | 313bp | 56 | |
| Suv39h2-qR | caaaatgagacacattgccgtat | ||||||
| G9a | 10919 | qRT-PCR | G9a-qF | ctaccgaacagccaagatg | 297bp | 61 | |
| G9a-qR | aactgaagaaggcgatgc | ||||||
| Phc2 | 1912 | qRT-PCR | Phc2-qF | aatcctgacgcatgttatcg | 275bp | 62 | |
| Phc2-qR | gcttggaacgcttgaacttat | ||||||
| Ring1 | 6015 | qRT-PCR | Ring1-qF | tgggaactgagtctgtatgagc | 229bp | 56 | |
| Ring1-qR | tctttcggcaggtaggaca | ||||||
| RD | ChIP-PS1 | RD-cF | ccacttatgcagttcctcacc | 216bp | 56 | NT008413.18 | |
| RD-cR | gtcattaaacaggctgaacc | ||||||
| p15 | 1030 | qRT-PCR | p15-qF | agtcaaccgtttcgggaggcg | 168bp | 62 | |
| p15-qR | accaccagcgtgtccaggaag | ||||||
| ChIP-PS2 | p15-cF | ggaacctagatcgccgatgtag | 74bp | 56 | |||
| p15-cR | tgttttacgcgtggaatgcac | ||||||
| p14-Arf | 1029 | qRT-PCR | p14-qF | gccaggggcgcccgccgctg | 236bp | 62 | |
| p14-qR | ggcccggtgcagcaccacca | ||||||
| ChIP-PS3 | p14-cF | gtgggtcccagtctgcagtta | 61bp | 56 | |||
| p14-cR | cctttggcaccagaggtgag | ||||||
| p16 | 1029 | (q)RT-PCR | p16-F | gctgcccaacgcaccgaata | 180bp | 62 | |
| p16-R | accaccagcgtgtccaggaa | ||||||
| ChIP-PS4 | p16-1cF | cggctgggagcagggaggc | 155bp | 62 | Bracken et al, 2007 | ||
| p16-1cR | gaatgtggcacccctgaagtcgc | ||||||
| ChIP-PS5 | p16-2cF | gtccccttgcctggaaagata | 154bp | 62 | Bracken et al, 2007 | ||
| p16-2cR | tctccgcagccgccgag | ||||||
| MSP-U | p16U-F | ttattagagggtggggtggattgt | 234bp | 62 | Herman et al, 1996 | ||
| p16U-R | ccacctaaatcaacctccaacca | ||||||
| MSP-M | p16M-F | ttattagagggtggggcggatcgc | 234bp | 62 | Herman et al, 1996 | ||
| p16M-R | ccacctaaatcgacctccgaccg | ||||||
| DHPLC | p16-uniF | tttttagaggatttgagggatagg | 392bp | 70→60 | Luo et al, 2006 | ||
| p16-uniR | ctacctaattccaattcccctacaaacttc | ||||||
| Col2A1 | Col2A1-qF | tctaacaattataaactccaaccaccaa | 92bp | 60 | Widschwendter et al, 2004 | ||
| Col2A1-qR | gggaagatgggatagaagggaatat | ||||||
| Col2A1-qProbe | ccttcattctaacccaatacctatcccacctctaaa | ||||||
| GAPDH | 2597 | (q)RT-PCR | GAPDH-F | gaaggtgaaggtcggagt | 226bp | 62 | |
| GAPDH-R | gaagatggtgatgggatttc |
DNA preparation, bisulfite treatment, and DNA methylation analysis
Genomic DNA of cell lines and tissue samples was isolated with phenol/chloroform extraction and modified with 5.0 M sodium bisulfite [33]. p16 methylation was analyzed with MSP, DHPLC, MethyLight, and clone sequencing [34]. Because of PCR bias favoring amplification of the unmethylated templates, the ratio (0.285) of the peak area for the methylated p16 to that for the unmethylated p16 in the p16-hemimethylated HCT116 cell was used as the constant to adjust the observed ratios for the tissue samples. Primer sets used were listed in Table 2.
RNA interference assay
The knockdown of Suv39h2 (sc-106822; Santa Cruz) and Suv39h1 (sc-38463; Santa Cruz) and G9a (sc-43777; Santa Cruz) was accomplished with siRNA duplex transiently. Scramble siRNA (5′-uucuc cgaac guguc acgu-3′) was used as the negative control. Expression level of target gene was analyzed with qRT-PCR and/or Western Blot assays 72 hours after transfection.
Chromatin Immunoprecipitation Assay (ChIP)
ChIP assays were performed and analyzed essentially as described [35]. The antibodies used were rabbit anti-H3K9me3 (07-625; Upstate), rabbit anti-H3K9me2 (07-441; Upstate), rabbit anit-H3K27me3 (07-449; Upstate), goat anti-SUV39H2 (Ab5264; Abcam), rabbit anti-Myc (Ab9132; Abcam). Rabbit anti-Myc antibody was used to precipitate exogenous CBX7 protein. The enrichment of specific genomic regions was assessed relative to the control IgG (ZB-2301, Zhongshan). Each ChIP experiment was in triplicates and repeated at least three times. Primer sets used were listed in Table 2.
Confocal Fluorescence Microscopy and BiFC assay
To produce fusion proteins for BiFC analysis, the N-terminal 172 amino acids of Venus (VN) was fused to the N-termini of human CBX7 protein (kindly provided by Claudius Vincenz) [23]. Amino acids 173–238 of YFP (YC, provided by Claudius Vincenz) were fused to the C-termini of SUV39H2. The sequences encoding all fusion proteins were verified and are available upon request. BiFC complexes and co-localization of different PcG proteins in transiently expressing cells were imaged 36 hours after transfection. Cells grown on cover glass were stained with 10 ng/ml Hoechst33342 (Sigma) for 30 min, and fixed in 2% formaldehyde for 10 min at room temperature, washed with phosphate-buffered saline, and mounted for microscopy. The fluorescence images were acquired by using a Leica inverted fluorescence microscope under oil objective. Blank VN- and YC-vector controls were used as negative controls. YC-Histone 3.1 (H3.1, kindly provided by Claudius Vincenz) was used as positive control.
Co-IP assay
After HEK293 were transfected with the YC-SUV39H2 (or -H3.1) and pcDNA3.1(+)/Myc-His-CBX7 vectors for 48 hours, proteins extracted were immunoprecipitated with mouse anti-Myc antibody (clone 9E10, Clontech) to precipitate exogenous CBX7 protein complexes and separated by SDS-PAGE and immunoblotted with a GFP antibody (ProteinTech Group). The mouse IgG antibody was used as negative control.
Supporting Information
DHPLC chromatogram of methylation status of p16 CpG island in human cancer cell lines.
(0.19 MB PDF)
The 3D images of chromodomain of the wildtype CBX7 (PDB id: 2K1B) and its mutant F11A, W32A produced by the DeepView software.
(0.29 MB PDF)
Distribution of CBX7-EGFP and CBX7-SUV39H2 complexes in human gastric carcinoma cell line MGC803.
(0.92 MB PDF)
Images of Co-IP assay for detection of CBX7-Histone 3.1 or CBX7-SUV39H2 complexes.
(0.09 MB PDF)
Effect of siRNA knockdown of HMTases G9a, SUV39H1 and SUV39H2 on formation of H3K9me3 within the p16 promoter and p16 expression in the CBX7 stably transfected subclone-2.
(0.20 MB PDF)
Effect of knockdown of Suv39h2 and Suv39h1 by siRNA on transcription of Suv39h2, Suv39h1, and p16 in PC3 cell line stably transfected with shRNA against Cbx7 or scramble shRNA control.
(0.46 MB PDF)
Acknowledgments
We are grateful to Claudius Vincenz for providing the VN-CBX7, YFP-CBX7, and YC-H3.1 vectors. We thank Kendra A. Williams and Shengyan Xiang for the proof reading of the manuscript. We also thank Dr. Yuanjia Chen, Dr. Guoren Deng, Dr. Xianzi Wen, Dr. Chengchao Shou, Dr. Qingyun Zhang, and Dr. Yang Ke for kindly sending us the tested cell lines.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work is supported by National Basic Research Program of China (973 Program 2005CB522403 and 2011CB504201) and Natural Science Foundation of China (30571056, 90919015, and 30921140311). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, et al. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 2.Gonzalez-Zulueta M, Bender CM, Yang AS, Nguyen T, Beart RW, et al. Methylation of the 5′ CpG island of the p16/CDKN2 tumor suppressor gene in normal and transformed human tissues correlates with gene silencing. Cancer Res. 1995;55:4531–4535. [PubMed] [Google Scholar]
- 3.Herman JG, Merlo A, Mao L, Lapidus RG, Issa JP, et al. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 4.Cao J, Zhou J, Gao Y, Gu L, Meng H, et al. Methylation of p16 CpG island associated with malignant progression of oral epithelial dysplasia: a prospective cohort study. Clin Cancer Res. 2009;15:5178–5183. doi: 10.1158/1078-0432.CCR-09-0580. [DOI] [PubMed] [Google Scholar]
- 5.Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- 6.Maertens GN, El Messaoudi-Aubert S, Racek T, Stock JK, Nicholls J, et al. Several distinct polycomb complexes regulate and co-localize on the INK4a tumor suppressor locus. PLoS One. 2009;4:e6380. doi: 10.1371/journal.pone.0006380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 8.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 9.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 10.Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, et al. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 13.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 14.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 15.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 16.Bernstein E, Duncan EM, Masui O, Gil J, Heard E, et al. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol. 2006;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Carroll D, Scherthan H, Peters AH, Opravil S, Haynes AR, et al. Isolation and characterization of Suv39h2, a second histone H3 methyltransferase gene that displays testis-specific expression. Mol Cell Biol. 2000;20:9423–9433. doi: 10.1128/mcb.20.24.9423-9433.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 19.Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gil J, Bernard D, Martinez D, Beach D. Polycomb CBX7 has a unifying role in cellular lifespan. Nat Cell Biol. 2004;6:67–72. doi: 10.1038/ncb1077. [DOI] [PubMed] [Google Scholar]
- 21.Bernard D, Martinez-Leal JF, Rizzo S, Martinez D, Hudson D, et al. CBX7 controls the growth of normal and tumor-derived prostate cells by repressing the Ink4a/Arf locus. Oncogene. 2005;24:5543–5551. doi: 10.1038/sj.onc.1208735. [DOI] [PubMed] [Google Scholar]
- 22.Gonzalez S, Klatt P, Delgado S, Conde E, Lopez-Rios F, et al. Oncogenic activity of Cdc6 through repression of the INK4/ARF locus. Nature. 2006;440:702–706. doi: 10.1038/nature04585. [DOI] [PubMed] [Google Scholar]
- 23.Vincenz C, Kerppola TK. Different polycomb group CBX family proteins associate with distinct regions of chromatin using nonhomologous protein sequences. Proc Natl Acad Sci U S A. 2008;105:16572–16577. doi: 10.1073/pnas.0805317105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Federico A, Pallante P, Bianco M, Ferraro A, Esposito F, et al. Chromobox protein homologue 7 protein, with decreased expression in human carcinomas, positively regulates E-cadherin expression by interacting with the histone deacetylase 2 protein. Cancer Res. 2009;69:7079–7087. doi: 10.1158/0008-5472.CAN-09-1542. [DOI] [PubMed] [Google Scholar]
- 25.Mohammad HP, Cai Y, McGarvey KM, Easwaran H, Van Neste L, et al. Polycomb CBX7 promotes initiation of heritable repression of genes frequently silenced with cancer-specific DNA hypermethylation. Cancer Res. 2009;69:6322–6330. doi: 10.1158/0008-5472.CAN-09-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang B, Pan K, Liu Z, Zhou J, Gu L, et al. Genetic polymorphisms of the E-cadherin promoter and risk of sporadic gastric carcinoma in Chinese populations. Cancer Epidemiol Biomarkers Prev. 2008;17:2402–2408. doi: 10.1158/1055-9965.EPI-08-0315. [DOI] [PubMed] [Google Scholar]
- 27.Dietrich N, Bracken AP, Trinh E, Schjerling CK, Koseki H, et al. Bypass of senescence by the polycomb group protein CBX8 through direct binding to the INK4A-ARF locus. Embo J. 2007;26:1637–1648. doi: 10.1038/sj.emboj.7601632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hahn MA, Hahn T, Lee DH, Esworthy RS, Kim BW, et al. Methylation of polycomb target genes in intestinal cancer is mediated by inflammation. Cancer Res. 2008;68:10280–10289. doi: 10.1158/0008-5472.CAN-08-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hinz S, Kempkensteffen C, Christoph F, Krause H, Schrader M, et al. Expression parameters of the polycomb group proteins BMI1, SUZ12, RING1 and CBX7 in urothelial carcinoma of the bladder and their prognostic relevance. Tumour Biol. 2008;29:323–329. doi: 10.1159/000170879. [DOI] [PubMed] [Google Scholar]
- 30.Pallante P, Federico A, Berlingieri MT, Bianco M, Ferraro A, et al. Loss of the CBX7 gene expression correlates with a highly malignant phenotype in thyroid cancer. Cancer Res. 2008;68:6770–6778. doi: 10.1158/0008-5472.CAN-08-0695. [DOI] [PubMed] [Google Scholar]
- 31.Scott CL, Gil J, Hernando E, Teruya-Feldstein J, Narita M, et al. Role of the chromobox protein CBX7 in lymphomagenesis. Proc Natl Acad Sci U S A. 2007;104:5389–5394. doi: 10.1073/pnas.0608721104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo WJ, Zeng MS, Yadav A, Song LB, Guo BH, et al. Mel-18 acts as a tumor suppressor by repressing Bmi-1 expression and down-regulating Akt activity in breast cancer cells. Cancer Res. 2007;67:5083–5089. doi: 10.1158/0008-5472.CAN-06-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eads C LP. DNA Methylation Protocols (Methods in Molecular Biology); 2002. Combined bisulfite restriction analysis (COBRA). pp. 53–70. [DOI] [PubMed] [Google Scholar]
- 34.Luo D, Zhang B, Lv L, Xiang S, Liu Y, et al. Methylation of CpG islands of p16 associated with progression of primary gastric carcinomas. Lab Invest. 2006;86:591–598. doi: 10.1038/labinvest.3700415. [DOI] [PubMed] [Google Scholar]
- 35.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DHPLC chromatogram of methylation status of p16 CpG island in human cancer cell lines.
(0.19 MB PDF)
The 3D images of chromodomain of the wildtype CBX7 (PDB id: 2K1B) and its mutant F11A, W32A produced by the DeepView software.
(0.29 MB PDF)
Distribution of CBX7-EGFP and CBX7-SUV39H2 complexes in human gastric carcinoma cell line MGC803.
(0.92 MB PDF)
Images of Co-IP assay for detection of CBX7-Histone 3.1 or CBX7-SUV39H2 complexes.
(0.09 MB PDF)
Effect of siRNA knockdown of HMTases G9a, SUV39H1 and SUV39H2 on formation of H3K9me3 within the p16 promoter and p16 expression in the CBX7 stably transfected subclone-2.
(0.20 MB PDF)
Effect of knockdown of Suv39h2 and Suv39h1 by siRNA on transcription of Suv39h2, Suv39h1, and p16 in PC3 cell line stably transfected with shRNA against Cbx7 or scramble shRNA control.
(0.46 MB PDF)