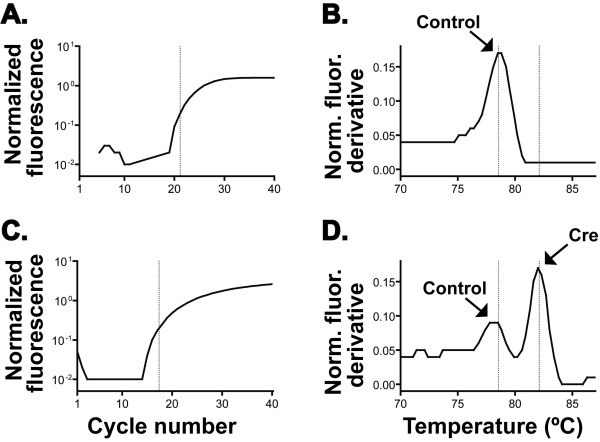
Figure 1.
Transgenic mouse genotyping using multiplex allele-specific PCR and melting curve analysis. PCR was performed in an optical cycler (ABI 7300) using 1-10 ng genomic DNA from mice of the indicated genotypes and a reagent mix containing SYBR GreenER. Amplification plots (A, C) and melting curves (B, D) are shown. Primers were designed to amplify 2 specific products: a genomic control product, generated from any genomic mouse template; and a transgene-specific product, generated only from genomic templates containing a cre transgene. Normalized fluorescence (y-axis, A &C) is the baseline-subtracted ratio of SYBR signal to ROX (passive reference dye) signal during amplification cycling (A, C). Normalized fluorescence derivative (y-axis, B & D) is the 2nd derivative of normalized fluorescence during the melting curve step. Dotted lines (A, C) indicate cycle threshold. A & B. Genomic DNA from a wild-type mouse; note robust amplification of the genomic control product with a single melting peak (allowing the distinction of a negative result from a failed PCR). C & D. Genomic DNA from a pdx1-creCre/0 mouse; note robust amplification with 2 distinct melting peaks corresponding to the control and cre-specific products. Arrows indicate presence of the genomic control product (Control) and the transgene-specific product (Cre).