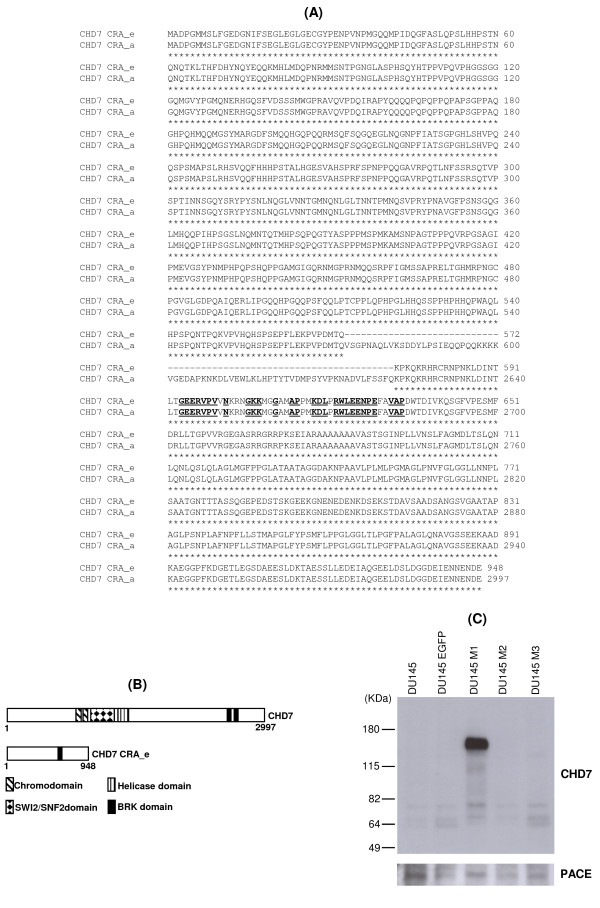
Figure 3.
Deduced protein sequence coded by the CHD7 CRA_e alternative transcript and respective Western blot analysis. (A) Alignment of the putative amino acids sequence coded by the CHD7 CRA_e alternative transcript with the amino acids sequence of the canonical CHD7 protein (accession number: NP_060250). The amino acids in bold and subtitled are conserved in the BRK domain (smart00592). The amino acids from the canonical CHD7 protein that are missing in the CHD7 CRA_e putative amino acid sequence were omitted for clarity (amino acids 601 to 2580). The numbers on the right refer to the amino acids at the end of each line related to each sequence. (B) Schematic representation of the domain structure of canonical CHD7 protein and CHD7 CRA_e isoform. (C) Western blot analysis of DU145 cells stably expressing CHD7 CRA_e alternative transcript (DU145 M1) using an anti-CHD7 antibody which recognizes the C-terminus end of CHD7. Parental DU145 cells and cells transduced with the empty vector (DU145 EGFP) or stably expressing two additional clones of CHD7 CRA_e alternative transcript, which putatively code for truncated proteins (DU145 M2 and DU145 M3) were used as negative controls. Detection of the PACE protein expression is shown as an internal protein loading control.