SYNOPSIS
Objective
Electronic health records (EHRs) have the potential to improve completeness and timeliness of tuberculosis (TB) surveillance relative to traditional reporting, particularly for culture-negative disease. We report on the development and validation of a TB detection algorithm for EHR data followed by implementation in a live surveillance and reporting system.
Methods
We used structured electronic data from an ambulatory practice in eastern Massachusetts to develop a screening algorithm aimed at achieving 100% sensitivity for confirmed active TB with the highest possible positive predictive value (PPV) for physician-suspected disease. We validated the algorithm in 16 years of retrospective electronic data and then implemented it in a real-time EHR-based surveillance system. We assessed PPV and the completeness of case capture relative to conventional reporting in 18 months of prospective surveillance.
Results
The final algorithm required a prescription for pyrazinamide, an International Classification of Diseases, Ninth Revision (ICD-9) code for TB and prescriptions for two antituberculous medications, or an ICD-9 code for TB and an order for a TB diagnostic test. During validation, this algorithm had a PPV of 84% (95% confidence interval 78, 88) for physician-suspected disease. One-third of confirmed cases were culture-negative. All false-positives were instances of latent TB. In 18 months of prospective EHR-based surveillance with this algorithm, seven additional cases of physician-suspected active TB were detected, including two patients with culture-negative disease. A review of state health department records revealed no cases missed by the algorithm.
Conclusions
Live, prospective TB surveillance using EHR data is feasible and promising.
The prevention and control of tuberculosis (TB) requires rapid and complete reporting of cases to health departments to facilitate contact tracing and assure timely therapy. The rise of multidrug-resistant TB with its attendant mortality has made timely surveillance more pressing than ever. Traditionally, health departments have relied on clinicians to report patients with TB, but this system has been associated with significant delays and underreporting.1,2 In the 1990s, the median reporting time for clinicians was up to 38 days, and as much as 18% of cases were not reported at all.3,4
During the past decade, automated laboratory reporting systems have improved the completeness and timeliness of reporting.5–7 These systems are blind, however, to cases of culture-negative disease. According to the Centers for Disease Control and Prevention (CDC), 13,299 cases of active TB were reported in the United States in 2007, and culture-negative TB accounted for 22% of these cases.8 In addition to missing cases of culture-negative disease, laboratory reporting systems typically do not report detailed patient demographics, clinician contact data, and prescribed medications.
Automated case identification and reporting from electronic health data is a promising new strategy to improve TB surveillance. Electronic health records (EHRs) include the same laboratory results that form the basis of automated laboratory reporting systems, as well as additional diagnostic, treatment, and demographic data. These data may be used to enhance disease detection and provide more complete information about cases to health officials.4,9–13 Detection using electronic health data lends itself well to automated electronic case reporting, because the data are continuously updated and often reside on Internet-enabled systems.
In the mid-1990s, Yokoe et al. grappled with the challenge of sensitive, accurate case detection using electronic insurance claims, pharmacy dispensing, and electronic medical record data from a health maintenance organization.4 They demonstrated that diagnosis codes alone have variable sensitivity and very poor positive predictive value (PPV) (presumably because similar codes can be used for both acute, active disease and old, healed disease). However, combinations of diagnostic codes, dispensed antituberculous medications, and procedure codes (e.g., chest radiographs or sputum staining for acid-fast bacteria) detected cases with high sensitivity, including many unreported cases. Dispensing of two or more antituberculous medications was the most sensitive criterion with a sensitivity of 89% (including many culture-negative cases), but with a PPV of only 30% secondary to false positives from nontuberculous mycobacteria.
Yokoe et al. reproduced these results in three different health plans across the country and called for development of a national surveillance system using this method.9 No such system has yet been developed, partly because many jurisdictions assign responsibility for reporting to clinicians rather than the insurers and pharmacy benefits management companies that are best positioned to link pharmacy-dispensing data to the patient and prescriber data required for effective reporting.
The recent development of EHR-based surveillance systems for real-time detection and reporting of notifiable diseases prompted us to adapt Yokoe et al.'s work to EHRs and assess the feasibility of real-time, prospective surveillance and reporting from EHR systems.10–13 Porting Yokoe et al.'s work to a live EHR-based surveillance environment required a change in the focus of TB algorithms from detection of confirmed disease to detection of suspected disease. This change is because definitive case confirmation often lags weeks to months after a case is first suspected due to the time required for clinical cultures to mature or to assess a patient's response to empiric therapy. However, public health officials typically prefer clinicians to report TB as soon as they suspect active disease rather than waiting for disease confirmation, so that prevention and control measures can be instituted immediately to prevent further spread of infection. If EHR-based surveillance systems are to complement or supersede traditional manual reporting, then electronic detection algorithms must be designed for timely reporting of suspected cases, despite knowing that many will ultimately turn out be something other than TB.
This article details the development of an EHR-specific TB detection algorithm and its implementation in a live, prospective surveillance and reporting system.
METHODS
Setting
All work was performed using the ambulatory EHR system of Atrius Health and its antecedent organizations (Harvard Vanguard Medical Associates and Harvard Community Health Plan). Atrius Health is a multi-specialty group practice with more than 700 physicians caring for approximately 600,000 adult and pediatric patients in 27 ambulatory care settings in eastern Massachusetts. All sites use the EpicCare EHR (Epic Systems, Verona, Wisconsin). The current system was introduced in 1997. Prior to this time, the practice used a noncommercial electronic medical record. At the time of conversion to EpicCare, both text and structured data were converted for most centers. EpicCare allows physicians to enter test orders and prescriptions, review test results, and assign diagnosis codes to each patient encounter. Prescription drugs are chosen from a searchable drug database arranged by both generic and trade names. Dispensing information is not recorded. Care delivered outside the practice, including most hospital care, is not coded, although scanned records are sometimes present in the electronic chart.
Algorithm development
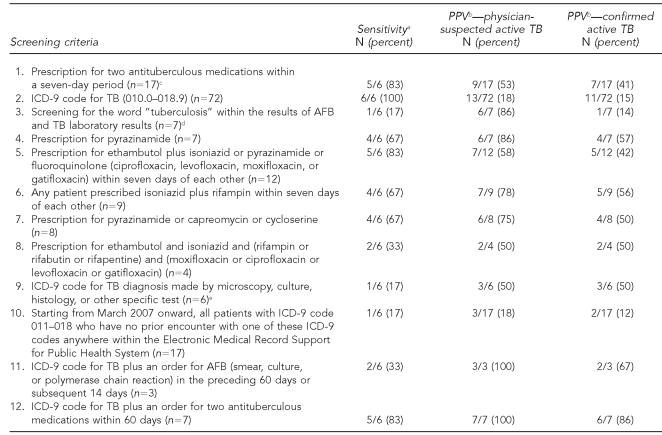
The creation of TB screening criteria was divided into development and validation steps. Algorithm development was conducted using EHR data spanning June 2006 to July 2007. We used clinical and research experience to develop 12 screening criteria to search for active TB (Table 1). These screening criteria incorporated the following EHR data elements: International Classification of Diseases, Ninth Revision (ICD-9) codes for TB, prescriptions for antituberculous medications, laboratory test orders and laboratory results for TB smears, cultures, and nucleic acid amplification tests. We did not incorporate automated pharmacy-dispensing data, such as were used by Yokoe et al., but instead relied on EHR prescription orders.
Table 1.
Candidate algorithms assessed for sensitivity and PPV for TB in a derivation cohort of electronic medical record data from Harvard Vanguard Medical Associates, June 2006 through July 2007
aReported relative to the total number of patients with confirmed active TB diagnosed or treated at a Harvard Vanguard Medical Associates facility.
bReported relative to the total number of patients captured by each individual algorithm. All captured cases of active TB were included when calculating PPV, including known patients diagnosed and treated at non-Harvard Vanguard Medical Associates facilities.
cList of included antituberculous medications: isoniazid, ethambutol, rifampin, rifabutin, rifapentine, pyrazinamide, streptomycin, para-aminosalicylic acid, kanamycin, capreomycin, cycloserine, and ethionamide
dScreened for Common Procedural Technology (CPT) 84460 (LxComponent 3836, 3838, 3840), CPT 87116 (LxComponent 2785, 3021, 3836, 3838, 3840, 4694, 931), CPT 87206 (LxComponent 3761), and CPT 87556 (LxComponent 4598)
eAny occurrence of 0.3–0.6 as the fourth digit of an ICD-9 code between 010 and 018
PPV = positive predictive value
TB = tuberculosis
ICD-9 = International Classification of Diseases, Ninth Revision
AFB = acid-fast bacilli
Each of the screening criteria was assessed for PPV and sensitivity. PPV for physician-suspected disease and confirmed active disease was determined by reviewing the full text medical record of each patient identified by the screening criteria. Each record was reviewed by two medical doctors (MC, MK); disagreements between reviewers were resolved by consensus. CDC's 1996 case definition was used as the reference standard.14 CDC's clinical case definition requires a positive tuberculin skin test, signs and symptoms consistent with TB (e.g., abnormal chest radiograph), treatment with at least two antituberculous medications, and an otherwise unrevealing diagnostic workup. The laboratory criteria require isolation of Mycobacterium tuberculosis (M. tuberculosis) from a clinical specimen, positive M. tuberculosis polymerase chain reaction, or the presence of acid-fast bacilli in a clinical specimen when cultures cannot be obtained. Cases that do not meet laboratory criteria for diagnosis of TB (culture-negative TB) require reevaluation after two months of empiric therapy to determine whether there has been a response to antituberculous therapy. If there is clinical or radiographic improvement in the absence of another diagnosis, the case is classified as active TB.15
We evaluated sensitivity by comparing the number of true cases of TB captured by the screening criteria with an independent list of all patients known to have active TB during the study period. This list was collated from (1) the study practice's infection control records, (2) a cross-match between all the practice's patients and the state health department's case list of TB cases diagnosed in Massachusetts during the study period, and (3) all confirmed cases of TB found by any of the screening criteria. Sensitivity is reported relative to the total number of patients with active TB diagnosed and/or treated within the practice. We excluded practice patients present in the state health department's list of confirmed cases if their TB was diagnosed and treated outside of the study practice (e.g., in a hospital or in the state's TB clinic).
Next, we selected the most promising screening criteria and combined them into a final algorithm with intent to achieve 100% sensitivity for confirmed active TB with the highest possible PPV for physician-suspected disease.
Algorithm validation using historical data
We validated the performance of the final algorithm using electronic medical record data spanning January 1990 through May 2006 (this includes some overlap with the population studied by Yokoe et al. during the years 1992 to 1996).4 During the validation phase, each medical record identified by the algorithm was abstracted by a nurse who determined whether a physician suspected active TB during evaluation of the patient and whether the case ultimately represented active TB, latent TB, or no TB. All abstractions were confirmed by a study physician (MC). Disagreements were resolved by consensus in consultation with an infectious disease specialist (MK). We were unable to evaluate sensitivity in the validation phase of the study due to the technical difficulty in matching the historical patient population with the state's TB archive.
Live, prospective surveillance
The final algorithm was then implemented into the Electronic Medical Record Support for Public Health (ESP) system.10–13 ESP is an EHR-based disease surveillance system designed to continually extract comprehensive encounter data from any source EHR, automatically apply disease detection algorithms to identify notifiable conditions, and then transmit electronic case reports to the state health department. ESP has been operational in Atrius Health since January 2007, extracting, analyzing, and reporting on new data every 24 hours. This ESP installation currently processes approximately 15,000 encounters, 20,000 laboratory results, and 6,000 prescription records per day. It reports cases of acute hepatitis A, B, and C; chlamydia; gonorrhea; pelvic inflammatory disease; and syphilis to the Massachusetts Department of Public Health (MDPH). ESP data extraction and data transmission modules can be modified by users to accept incoming data from different EHR systems and to transmit case reports in custom formats. Further documentation and free source codes for ESP are available under a lesser general public-use agreement at .www.esphealth.org We assessed PPV and the completeness of case capture relative to conventional reporting in 18 months of prospective surveillance within ESP.
RESULTS
Algorithm development
All candidate screening criteria are presented in Table 1. Six patients were diagnosed and/or treated for active TB from June 2006 through July 2007. These included two patients with culture-negative disease. Candidate screening criteria captured between one and six of these patients. The PPVs of various criteria for physician-suspected active TB ranged from 18% to 100%.
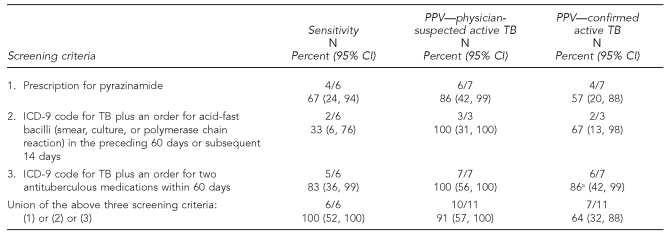
Three of the candidate screening criteria were combined into a final algorithm: (1) prescription for a medication regimen including pyrazinamide, or (2) ICD-9 code for TB plus an order for acid-fast bacilli (smear, culture, or polymerase chain reaction) in the preceding 60 days or subsequent 14 days, or (3) ICD-9 code for TB plus an order for at least two antituberculous medications other than pyrazinamide within 60 days (Table 2). The final algorithm detected 11 patients, of whom 10 had physician-suspected active TB (PPV=91%). Of these, seven were ultimately confirmed to have active disease (PPV=64%). These included all six cases with TB diagnosed or treated in the practice between June 2006 and July 2007, as well as one additional case diagnosed and treated outside of the practice. The one false-positive patient without suspicion of active disease had latent TB. The algorithm captured the patient after an erroneous prescription for pyrazinamide (canceled by the physician within a day of prescribing).
Table 2.
Sensitivity and PPV of a final algorithm for detection of physician-suspected TB using electronic medical record data, Harvard Vanguard Medical Associates, June 2006 through July 2007
aThis algorithm picked up one patient who was diagnosed and treated at an outside hospital, accounting for the different numerator when calculating PPV.
PPV = positive predictive value
TB = tuberculosis
CI = confidence interval
ICD-9 = International Classification of Diseases, Ninth Revision
Of the 10 physician-suspected cases detected by our final algorithm, seven had been reported to the health department for suspected disease. The three patients hitherto unknown to the health department included one patient with culture-negative disease. The other two cases were physician-suspected cases that were ultimately diagnosed with other conditions.
Algorithm validation using historical data
The final algorithm was validated in electronic data spanning January 1990 through May 2006. The algorithm identified 218 patients, of whom the treating physician suspected active TB in 183 (84%). The remaining 35 were cases of latent TB.
Of the 183 cases of suspected active TB, 103 were ultimately confirmed (56%). Hence, the final algorithm's PPV was 84% for suspected active TB (95% CI 78, 88) and 47% for confirmed active TB (95% CI 41, 54). A total of 33 of the 103 cases of confirmed active TB (32%) were culture-negative disease, with 67% being cases of pulmonary TB and 33% being cases of extra-pulmonary TB. This breakdown was similar in the cases of culture-positive disease, with 63% being cases of pulmonary TB and 37% being cases of extra-pulmonary TB. Patients ranged in age from one to 85 years and 55% were female (data not shown).
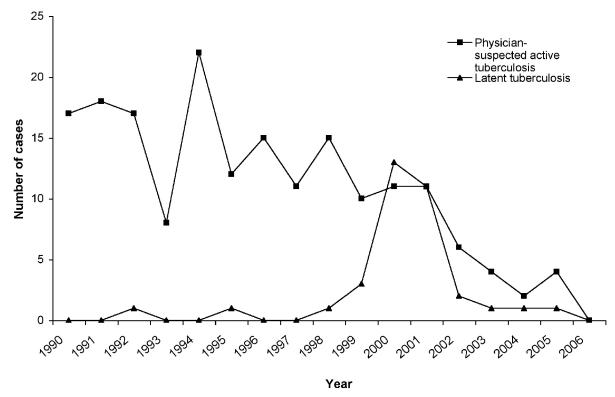
The number of cases detected each year and their breakdown into physician-suspected active TB and latent TB is shown in the Figure. The algorithm identified a mean of 16.4 cases per year through 2001, and then a mean of 5.3 cases per year from 2002 to 2005 (last complete year). Notably, 26 of the 35 instances of latent TB occurred between 1999 and 2002. No more than one case per year of latent disease was found before 1999 or after 2002.
Algorithm validation in live, prospective surveillance
The final algorithm was implemented in the ESP system in August 2007.10–13 In 18 months of prospective, live surveillance from August 2007 to January 2009, ESP detected seven additional cases of TB and electronically reported them to MDPH. All cases were physician-suspected disease. Six of the seven cases were eventually confirmed to be active TB, while the seventh was revoked after further investigation. Two of the six confirmed cases were culture-negative disease. Review of state health department records of TB cases reported by any source has not revealed any patients from the study practice missed by ESP.
All cases were initially diagnosed in hospitals (i.e., outside the ambulatory practices covered by ESP) and reported by the hospitals to the state health department via conventional reporting methods. Despite the hospitals' lead-time advantage, however, ESP detected two of the cases before the hospitals reported to the health department (12 days and 36 days, respectively, before manual reporting). All other cases were detected by ESP after they had already been reported by the diagnosing hospitals.
DISCUSSION
An algorithm incorporating medication prescriptions, diagnosis codes, and laboratory orders accurately detected physician-suspected TB when applied to EHR data. In 18 months of live, prospective surveillance, the algorithm detected seven cases with a PPV of 100% for physician-suspected active TB and no known missed cases of confirmed active TB (sensitivity 100%). Although these numbers are small and warrant guarded interpretation, they are consistent with the accuracy of the algorithm during development with data from 2006 to 2007 (sensitivity 100%, PPV for physician-suspected disease 91%) and during validation with 18 years of historical data spanning 1990–2006 (sensitivity not available, PPV for physician-suspected disease 84%). There were no false-positive patients without any connection to TB. The only patients detected without confirmed active disease either had physician-suspected disease (and, hence, should have been reported by conventional surveillance) or latent TB (a reportable condition in Massachusetts, although not reportable in all states). Approximately one-third of confirmed cases detected by the algorithm were instances of culture-negative disease. These were high-value cases because, by definition, they were cases that were missed by laboratory-based surveillance systems.
Evaluation of the relative timeliness of EHR-based reporting vs. conventional reporting (laboratory- and clinician-based) mechanisms was limited in this study because all cases detected during live, prospective surveillance were first diagnosed outside of the ambulatory system covered by our electronic surveillance system. Not surprisingly, the majority of patients were therefore reported to the health department by laboratory systems and outside facility clinicians first. Notably, however, there were two patients who were electronically detected in our ambulatory practice surveillance system before the outside hospitals submitted case reports, despite their lead time in making the initial diagnosis. This lag time suggests that broader deployment of EHR-based surveillance systems to cover inpatient as well as outpatient systems may substantially improve the timeliness of reporting for all cases.
The rate of false-positive cases found by the final algorithm varied across time. Notably, 26 of the 35 (74%) cases of latent TB occurred between 1999 and 2002 (Figure). This period coincides with former CDC and American Thoracic Society (ATS) guidelines for treatment of latent TB that included an option for two-month therapy with rifampin/pyrazinamide.16 In 2003, however, CDC/ATS cautioned against the use of pyrazinamide to treat latent TB due to excessive hepatotoxicity.15 Following 2003, the number of latent TB cases found by the algorithm markedly decreased as clinicians stopped prescribing pyrazinamide for latent TB but instead reserved it as a core part of multidrug regimens for active TB. Pyrazinamide is not recommended for treatment of nontuberculous mycobacteria or for any other medical conditions. Consequently, since 2003, the PPV of the algorithm has been 87% for physician-suspected disease and 65% for confirmed active disease. As such, the algorithm is optimized for contemporary treatment recommendations. Future changes to treatment recommendations, particularly concerning the use of pyrazinamide, could substantially change the performance of the algorithm and necessitate modifications to maintain accuracy.
Figure.
Number of cases of physician-suspected active tuberculosis and latent tuberculosis detected by the final algorithm with the validation cohort of electronic medical record data, Harvard Vanguard Medical Associates, 1990–2006a
aThe marked increase in latent tuberculosis (TB) cases detected by the final algorithm between 1999 and 2002 coincides with former Centers for Disease Control and Prevention/American Thoracic Society guidelines for the treatment of latent TB that included an option for two-month therapy with rifampin/pyrazinamide. The data from 2006 are only reported through May.
The percentage of electronically suspected cases confirmed to have active TB (56%, 95% CI 49, 63) mirrors the percentage of conventionally reported cases confirmed to have active disease. In 2007, for example, the MDPH received 533 reports of suspected TB cases and subsequently confirmed active TB in 248 (46%) of those cases (Personal communication, Jessica Malenfant, MDPH, October 2007). As previously mentioned, public health officials encourage early reporting of suspected cases to assure rapid and sensitive surveillance. Our algorithm's high PPV for suspected disease at the cost of moderate PPV for confirmed disease mirrors this philosophy.
Limitations
It is possible that the algorithm did miss some cases of true disease. We were not able to validate its sensitivity in retrospective data due to a technical inability to cross-match the medical practice's historical patient population with the state's records of TB patients. During derivation and live implementation, however, comparison with state health department records did not reveal any patients missed by the algorithm. Rather, the converse was true. The algorithm uncovered some patients with confirmed disease who had not been reported to the health department via conventional channels. The decline in suspected and reported cases from 2002 to present might also suggest missing cases; however, this trend mirrors the steady decrease in TB incidence reported nationally after a surge in the 1990s.8
The construction of this algorithm evolved out of the work by Yokoe et al. in the 1990s.4,9 Yokoe et al. found that dispensing at least two antituberculous medications was the most promising criteria for detecting active TB. Our final algorithm included a variation of this rule, namely a prescription for (rather than dispensing of) at least two antituberculous medications and an ICD-9 code for TB. Requiring the presence of an ICD-9 code along with the prescription for antituberculous medications increased the PPV for TB by decreasing false positives due to nontuberculous mycobacteria. Likewise, we added two more criteria in our final algorithm: a prescription for pyrazinamide or the combination of a lab order and an ICD-9 code for TB. These options were added to capture the few cases missed by screening for at least two antituberculous medications alone.
Broader adoption of EHR-based TB surveillance faces many barriers. Foremost among these is the low rate of EHR adoption by U.S. physicians. At present, only 17% of U.S. doctors have even basic electronic medical record systems.17,18 There are, however, many local and national initiatives to promote greater adoption of health information technology and to facilitate EHR use for public health needs. The Health Information Technology Standards Panel (http://www.hitsp.org) recently published specifications for standardized Public Health Care Reporting from EHRs.19 The algorithm developed in this study is a model for how EHRs can automatically identify cases that can then be reported using these emerging standards. Now is the time to begin preparing the public health landscape to take advantage of the improved capacity for surveillance that may be possible as more health-care providers adopt EHRs.
CONCLUSIONS
This work demonstrates the feasibility of real-time, prospective surveillance of EHR data for active TB. We demonstrated that it is possible to perform automated, prospective detection and reporting of active TB cases by using ambulatory EHR data that contain diagnosis, laboratory test, and prescribing information. This approach to surveillance is especially important to identify the substantial fraction of active cases that are culture-negative, to permit early notification of cases at the point of physician suspicion, and to convey complete contact information about patients and the diagnosing clinician. Broad deployment of such systems has the potential to substantively improve the comprehensiveness, efficiency, and timeliness of TB surveillance.
Acknowledgments
The authors thank Victoria J. Morrison, RN, PhD (Salem State College, Sale, Massachusetts), for her assistance in chart abstraction, and Deborah S. Yokoe, MD, MPH (Brigham and Women's Hospital and Harvard Medical School, Boston, Massachusetts), for her thoughtful review of this article.
Footnotes
This work was supported by Centers for Disease Control and Prevention grant #8P011HK000016-04.
REFERENCES
- 1.Doyle TJ, Glynn MK, Groseclose SL. Completeness of notifiable infectious disease reporting in the United States: an analytical literature review. Am J Epidemiol. 2002;155:866–74. doi: 10.1093/aje/155.9.866. [DOI] [PubMed] [Google Scholar]
- 2.Jajosky RA, Groseclose SL. Evaluation of reporting timeliness of public health surveillance systems for infectious diseases. BMC Public Health. 2004;4:29. doi: 10.1186/1471-2458-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Curtis AB, McCray E, McKenna M, Onorato IM. Completeness and timeliness of tuberculosis case reporting. A multistate study. Am J Prev Med. 2001;20:108–12. doi: 10.1016/s0749-3797(00)00284-1. [DOI] [PubMed] [Google Scholar]
- 4.Yokoe DS, Subramanyan GS, Nardell E, Sharnprapai S, McCray E, Platt R. Supplementing tuberculosis surveillance with automated data from health maintenance organizations. Emerg Infect Dis. 1999;5:779–87. doi: 10.3201/eid0506.9906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Effler P, Ching-Lee M, Bogard A, Ieong MC, Nekomoto T, Jernigan D. Statewide system of electronic notifiable disease reporting from clinical laboratories: comparing automated reporting with conventional methods. JAMA. 1999;282:1845–50. doi: 10.1001/jama.282.19.1845. [DOI] [PubMed] [Google Scholar]
- 6.Panackal AA, M'ikanatha NM, Tsui FC, McMahon J, Wagner MM, Dixon BW, et al. Automatic electronic laboratory-based reporting of notifiable infectious diseases at a large health system. Emerg Infect Dis. 2002;8:685–91. doi: 10.3201/eid0807.010493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Overhage JM, Grannis S, McDonald CJ. A comparison of the completeness and timeliness of automated electronic laboratory reporting and spontaneous reporting of notifiable conditions. Am J Public Health. 2008;98:344–50. doi: 10.2105/AJPH.2006.092700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention (US) Reported tuberculosis in the United States, 2007. Atlanta: CDC; 2008. [Google Scholar]
- 9.Yokoe DS, Coon SW, Dokholyan R, Iannuzzi MC, Jones TF, Meredith S, et al. Pharmacy data for tuberculosis surveillance and assessment of patient management. Emerg Infect Dis. 2004;10:1426–31. doi: 10.3201/eid1008.031075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klompas M, Lazarus R, Daniel J, Haney GA, Campion FX, Kruskal BA, et al. Electronic Medical Record Support for Public Health (ESP): automated detection and reporting of statutory notifiable diseases to public health authorities. Adv Dis Surveill. 2007;3:3. [Google Scholar]
- 11.Automated detection and reporting of notifiable diseases using electronic medical records versus passive surveillance—Massachusetts June 2006-July 2007. MMWR Morb Mortal Wkly Rep. 2008;57(14):373–6. [PubMed] [Google Scholar]
- 12.Klompas M, Haney G, Church D, Lazarus R, Hou X, Platt R. Automated identification of acute hepatitis B using electronic medical record data to facilitate public health surveillance. PLoS ONE. 2008;3:e2626. doi: 10.1371/journal.pone.0002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazarus R, Klompas M, Campion FX, McNabb SJN, Hou X, Daniel J, et al. Electronic support for public health: validated case finding and reporting for notifiable diseases using electronic medical data. J Am Med Inform Assoc. 2009;16:18–24. doi: 10.1197/jamia.M2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Centers for Disease Control and Prevention (US) Case definitions for infectious conditions under public health surveillance—tuberculosis. [cited 2008 Dec 30]. Available from: URL: http://www.cdc.gov/ncphi/disss/nndss/casedef/tuberculosis_current.htm.
- 15.Blumberg HM, Burman WJ, Chaisson RE, Daley CL, Etkind SC, Friedman LN, et al. American Thoracic Society/Centers for Disease Control and Prevention/Infectious Diseases Society of America: treatment of tuberculosis. Am J Respir Crit Care Med. 2003;167:603–62. doi: 10.1164/rccm.167.4.603. [DOI] [PubMed] [Google Scholar]
- 16.American Thoracic Society. Targeted tuberculin testing and treatment of latent tuberculosis infection. Am J Respir Crit Care Med. 2000;161(4 Pt 2):S221–47. doi: 10.1164/ajrccm.161.supplement_3.ats600. [DOI] [PubMed] [Google Scholar]
- 17.Blumenthal D. Stimulating the adoption of health information technology. N Engl J Med. 2009;360:1477–9. doi: 10.1056/NEJMp0901592. [DOI] [PubMed] [Google Scholar]
- 18.The National Alliance for Health Information Technology. Report to the Office of the National Coordinator for Health Information Technology on defining key health information technology terms. Rockville (MD): Department of Health and Human Services (US); 2008 Apr 28; [cited 2010 Jan 9]. Also available from: URL: http://healthit.hhs.gov/portal/server.pt/gateway/PTARGS_0_10741_848133_0_0_18/10_2_hit_terms.pdf. [Google Scholar]
- 19.Healthcare Information Technology Standards Panel. IS 11—public health case reporting. [cited 2010 May 23]. Available from: URL: http://www.hitsp.org/InteroperabilitySet_Details.aspx?MasterIS=true&InteroperabilityId=364&PrefixAlpha=1&APrefix=IS&PrefixNumeric=11.