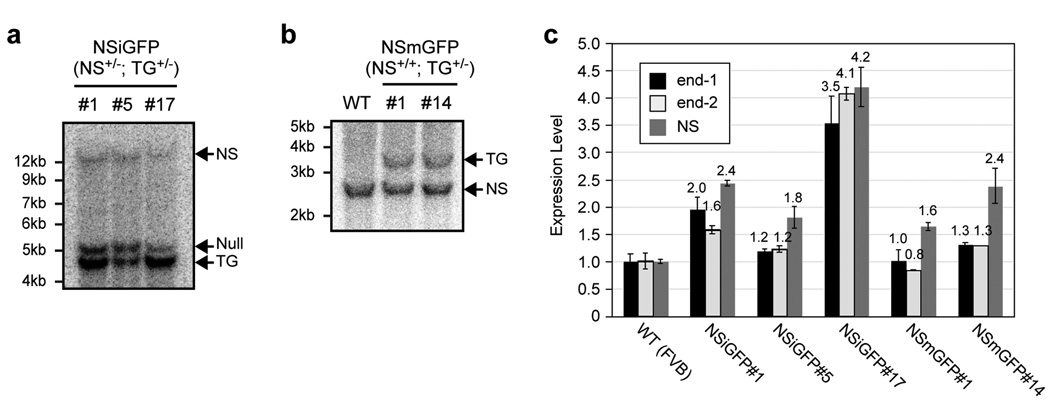
Figure 2. Measurement of the transgene copy number and integrity in the NSiGFP and NSmGFP lines.
(a) Three NSiGFP lines were established and mated with heterozygous NS+/− mice. Their offspring (NS+/−; NSiGFP+/−) were genotyped by BamHI-digested Southern blots hybridized with the right-arm probe indicated in Fig. 1a, where the nucleostemin transgene (TG), the nucleostemin-null (null), and the endogenous nucleostemin alleles (NS) are represented by the 4.6kb, 5.2kb, and 14.3kb fragments, respectively. Images were scanned and digitally quantified using the ImageJ 1.36b software. The transgene copy number is determined to be 2, 1, and 4 for NSiGFP#1, #5, and #17 lines, based on the intensity ratio between the TG and the null allele. (b) Two NSmGFP lines (#1 and #14) were established. Their transgene copy numbers are both determined to be one based on the intensity ratio between the TG (3.6 kb) and the NS fragments (2.6kb) in the (NS+/+; TG+/−) mice. Southern blots were digested with Bgl2 and hybridized with the left-arm probe. (c) The integrity of the BAC transgene was assessed by qPCR assays using primers that recognized the BAC ends (end-1 and end-2) and the middle portion of the nucleostemin gene (NS). Our data indicates that the end of the transgene is better preserved in the multi-copy lines (NSiGFP#1 and #17) than in the single-copy lines (NSiGFP#5, NSmGFP#1, and NSmGFP#14).