Abstract
The multicopy λgt10-lacZ transgene shuttle vector of Muta™Mouse serves as an important tool for genotoxicity studies. Here, we describe a model for λgt10-lacZ transgene molecular structure, based on characterisation of transgenes recovered from animals of our intramural breeding colony. Unique nucleotide sequences of the 47 513 bp monomer are reported with GenBank® assigned accession numbers. Besides defining ancestral mutations of the λgt10 used to construct the transgene and the Muta™Mouse precursor (strain 40.6), we validated the sequence integrity of key λ genes needed for the Escherichia coli host-based mutation reporting assay. Using three polymerase chain reaction (PCR)-based chromosome scanning and cloning strategies, we found five distinct in vivo transgene rearrangements, which were common to both sexes, and involved copy fusions generating ∼10 defective copies per haplotype. The transgene haplotype was estimated by Southern hybridisation and real-time–polymerase chain reaction, which yielded 29.0 ± 4.0 copies based on spleen DNA of Muta™Mouse, and a reconstructed CD2F1 genome with variable λgt10-lacZ copies. Similar analysis of commercially prepared spleen DNA from Big Blue® mouse yielded a haplotype of 23.5 ± 3.1 copies. The latter DNA is used in calibrating a commercial in vitro packaging kit for E.coli host-based mutation assays of both transgenic systems. The model for λgt10-lacZ transgene organisation, and the PCR-based methods for assessing copy number, integrity and rearrangements, potentially extends the use of Muta™Mouse construct for direct, genomic-type assays that detect the effects of clastogens and aneugens, without depending on an E.coli host, for reporting effects.
Introduction
Knowledge concerning spontaneous and induced mutagenesis in somatic and germ line tissues has been greatly enhanced by use of transgenic rodent systems such as Muta™Mouse (1,2) and BigBlue® mouse (3,4) and related cell lines (5–7). These transgenes are composed of engineered λ-bacteriophage and mutation reporter genes from Escherichia coli (lacZ for Muta™Mouse and lac I for BigBlue®). The λ-components are essential for rescue of the reporter genes from the murine genome, and their delivery, amplification and detection (plaque assay) using select E.coli hosts. The implicit assumptions in the use of these mutation reporting systems are that all the transgenic λ-shuttle vector copies are identical, and can be efficiently and independently recovered from genomic DNA of any cell or tissue type of either sex, and are expressible in E.coli. However, the nucleotide sequence of either transgene system has not been verified, including the λ-related precursor components as well as any potential alterations arising from the actual integration and the formation of multiple copies of the transgenes during establishment of the mouse lines (1,3,8–10). Estimates of mutation frequencies of the E.coli reporter gene(s) may not accurately reflect those of endogenous loci (11). Features that distinguish these transgenes (e.g. λgt10-lacZ) from most endogenous genes include: their multicopy, head-to-tail chromosomal arrangements, their extensive and high CpG content relative to the mouse genome, methylation, phasing in replication and lack of transcription (i.e. no transcripts detected in RNA from kidney, liver and lung as reported by [(1,12) and Shwed, P. unpublished results]. Several of these properties are not well understood and some may be disruptive to chromosome-chromatin domain structure roles such as transcription, replication, recombination and repair (13–17). Furthermore, transgene reporters may not be recovered or expressed in E.coli hosts because of mutations that disable λ-genes and/or generate sequence conversions for prophage-like insertions (18).
To better understand the integrity and chromosomal structure of Muta™mouse λgt10-lacZ copies, we undertook a characterisation of the monomer sequence in animals from our colony. Here, we report the novel nucleotide sequence data, providing information on the transgene’s ancestry with respect to λgt10 and its two precursor lambdoid genomes and integrity of the transgene copies in tissues. As part of the study, the transgene copy number was investigated by DNA hybridisation (Southern blot) and quantitative real-time–polymerase chain reaction (RT–PCR), using NCBI mouse genome build 36.1 to derive copy reference standards such as single copy and multicopy endogenous genes or mixtures of λgt10-lacZ transgene and mouse genomic DNA. Further, we included a copy comparison with commercially available DNA from BigBlue® mouse, which could serve as a potential inter-laboratory reference standard.
Materials and methods
Sources of genomic DNA, transgene and standards
Muta™Mouse strain 40.6 was obtained directly from the Jan Vijg laboratory (TNO, The Netherlands) in 1990 (2) and thereafter maintained as a breeding colony under approval of Health Canada’s Animal Care Committee. Information on the strain’s construction and protocols for isolation of high-molecular weight DNA from various tissues are reported elsewhere (2,19).
Additional Muta™Mouse liver tissue was obtained from Covance Laboratories Ltd, Harrogate, UK. Monomers of λgt10-lacZ were obtained from Muta™Mouse DNA using Transpack® packaging extract (Stratagene, La Jolla, CA, USA) and a Wizard® Lambda Preps DNA Purification Kit (Promega, Madison, WI, USA), according to manufacturer’s protocols. Cloning vector λgt10 and λC1857 were obtained from Sigma. Standards to determine copy number of λgt10-lacZ DNA per diploid genome were made by mixing 1 μg of non-transgenic mouse CD2F1 [(BALB/c × DBA/2) F1] spleen DNA (Jackson Laboratory, Bar Harbor, ME, USA) with λgt10-lacZ DNA at various concentrations. Copy number was based on NCBI mouse genome build 36.1 (5.1 × 109 bp per haploid cell) and the 47 513 bp λgt10-lacZ model (see Figure1). BigBlue® mouse spleen DNA was supplied with the Transpack® packaging kit. DNA quality (260/280 nm and 260/230 nm) and quantity (A260 nm) parameters were obtained by using the ND 1000 micro spectrophotometer (Nano Drop Technologies, Wilmington, DE, USA).
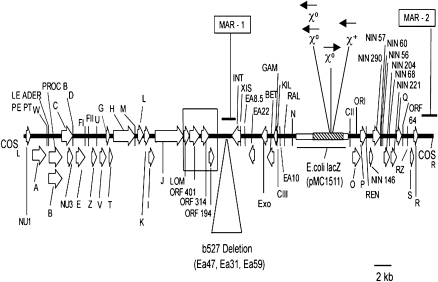
Fig. 1.
Model of the Muta™Mouse λgt10-lacZ transgene derived by sequence analysis. The transgene monomer has 47 513 bp and 57 ORFs and is based on nucleotide sequencing of PCR amplicons derived from systematic scanning of functional regions of λgt10-lacZ using Muta™Mouse genomic DNA from tissues of both gender and λgt10-lacZ in vivo copies rescued by in vitro phage packaging and commercial stocks of λCI857 and λgt10. Data include the b527 deletion and unfinished parts of bacteriophage imm434 (accession numbers M60848, Y00118) and the 5.2 kb EcoR1-DraI fragment of pMC1511, which contains the E.coli lacZ mutation reporter (GenBank® L08935) (hatched-box). The GenBank® accession numbers for sequences other than those reported already for λ (NC_001416; 48.5 kb) are given in Table II. Functional regions included left (COSL) and right (COSR) cohesive ends and ORFs required for virion assembly and DNA packaging and origin of λ DNA replication (ORI). Arrows show orientation of ORFs common to the 48.5 kb λ bacteriophage as identified by λ gene nomenclature (see NC_001416). Novel in vivo copy rearrangements are described in Figure 2 and Table II. The large open box spans a novel finding of a region of substitution by lambdoid Rac prophage tail fiber assembly gene (lom and ORFs 401, 314 and 194) found also in λgt10 and λCI857 commercial stocks. Also identified are crossover hot spot instigator motifs (Chi sites) conveying potential for lacZ recombination with the E.coli genome. The symbol χ+ signifies a fully functional Chi motif (starts at nucleotide position 3508 on the reverse or antistrand with respect to E.coli lacZ (accession J01636). χ0 signifies a one base difference version of the Chi motif that requires a single mutation for full function. These χ0sites start at nucleotide positions 3149 and 3152 for the two on the anti-strand and position 3248 for the one on the sense strand. MAR-1 and MAR-2 show location of sequences identified as matrix-associated regions using a computational approach (15). MAR-1 is located 5′ to the lambda INT gene and MAR-2 is located in the 3′ non-coding λgt-10 region and they represent regions that may contribute to higher order mammalian chromosome loop structures. In a head-to-tail arrangement of transgenes, MAR-2 would be located within 1.4 kb of the COSL site and in the vicinity of rearrangements featured in Figure 2.
Sequence analysis and derivation of DNA probes
Unless otherwise indicated, most of the DNA sequences used and generated in this work are identified by GenBank® accession numbers (see Tables I and II, and legends to Figures 1–4). The oligonucleotide primers (see list in Table I) were designed using Vector NTI™ version 9.0.0 software (Invitrogen, Burlington, Ontario, Canada) and GenBank® sequence data for λ bacteriophage, imm434 and E.coli lacZ. These custom-made primers (Cortec DNA Service Laboratories Inc., Kingston, Canada) were used to generate polymerase chain reaction (PCR) fragments in reactions involving a DNA polymerase deficient in 3′–5′ exonuclease activity (KOD polymerase; Novagen, Gibbstown, NJ, USA) to minimise proofreading errors. Fragments generated, either by PCR or cloning in E.coli, were sequenced in both directions using an ABI Prism® 3100 Genetic Analyzer and BigDye® Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). The nucleotide sequences were compiled with Vector NTI software and communicated to GenBank® using its software (Bankit).
Table I.
Primers used in this study for PCR and nucleotide sequencing
| Gene target/amplicon size (bp) | Primer pair (forward/reverse) |
| General sequencing primers | |
| λNU region (600 bp) | Nu1-1: 5′-CGGGTTTTCGCTATTTATGA-3′ |
| Nu1-2: 5′-ACATGTCGGTTTTCCAGTTC-3′ | |
| λlom, ORF401-like coding sequence side tail fiber and INT genes (3929 bp) | LO-1: 5′-ATGGCAGTAAAGATTTCAGG-3′ |
| LO-2: 5′-ACAGCAGGCCACTCAATATC-3′ | |
| λb527 mutation (972 bp) | B5-1: 5′-GAATCAATTCCAATTACCTGAAGTC-3′ |
| B5-2: 5′-GTATCAAGGTATTTTATGCGC-3′ | |
| λN-O region (6551 bp) | NO-1 5′-TTTTCCCTTAATTTTCTGGC-3′ |
| NO-2: 5′-TTACCGGACCAGAAGTTCTC-3′ | |
| E.coli lacZ (3341 bp) | LZ-1: 5′-GAATTCCGATTCATTAAT-3′ |
| LZ-2: 5′-GAATTCAAATAGTACATAATGG-3′ | |
| λTail region (582 bp) | TR-1: 5′-TATGTCCACAGCCCTGACG-3′ |
| TR-2: 5′-CTCGTATCACATGGAAGG-3′ | |
| λReplication origin (ori) (149 bp) (RT-PCR) | ORI-1: 5′-GACCCAACTCGAAATCAAC-3′ |
| ORI-2: 5′-ATCTGCTCACGGTCAAAG-3′ | |
| (501 bp) Ori Southern hybridization | ORI-S1: 5′-GGACAGGAGCGTAATGTGGCA-3′ |
| ORI-S2: 5′-AATTGCAGCATCCGGTTTCAC-3′ | |
| (812 bp) 18S Southern hybridization | 18SS-1: 5′-TCTTTCGAGGCCCTGTAAT-3′ |
| 18SS-2: 5′-ACCAACTAAGAACGGCCAT-3′ | |
| Mus musculus 18S rDNA gene (117 bp) (RT-PCR) | 18S-1: 5′-ACGGACAGGATTGACAGATTG-3′ |
| 18S-2: 5′-CCAGAGTCTCGTTCGTTATCG-3′ | |
| Mus musculus annexin V gene exon 4 (115 bp) (RT-PCR) | AN-1: 5′-GCATCCTGAACCTGTTGACATC-3′ |
| AN-2: 5′-ACACCCACTCCACCTTGAATG-3′ | |
| λ inverse primers | |
| (Position 340) | INV-1: 5′-CTTTTTGGCCTCTGTCGTT-3′ |
| (Position 97) | INV-2: 5′-TCCTTTCTTTTCAGAGGGTA-3′ |
| Genome walking λgt10-lacZ primers | |
| (Extending into 5′end, position 101) | GW-1 5′-CACCTGTCGTTTCCTTTCTTTTCAGAG- 3′ |
| (Extending into 5′end, position 44) | GW-2: 5′-GAAGAAGAACGGAAACGCCTTAAACC-3′ |
| (Extending to 3′ end, position 48195) | GW-3: 5′-AATTCCCGGACCCTTTTTGCTCAAGAGC-3′ |
| (Extending to 3′end, position 48475) | GW-4: 5′-GTCCTTTCCGGTGATCCGACAGGTTAC-3′ |
| Primers to scan for detection of transgene integration | |
| Biotinylated transgene primer | BIO-1 5′-CAGCATCCCTTTCGGCATACCATT-3′ |
| Degenerate primers | |
| FP1 | 5′-GACTCAGATATCGGCAGCGTGGTNNNNNNNGCGCT-3′ |
| FP2 | 5′-GACTCAGATATCGGCAGCGTGGTNNNNNNNGGCCT-3′ |
| FP3 | 5′-GACTCAGATATCGGCAGCGTGGTNNNNNNNGCGCA-3′ |
| FP4 | 5′-GACTCAGATATCGGCAGCGTGGTNNNNNNNGGCCA-3′ |
| Nested gene specific primers | |
| GSP1 | 5′-ACTATAGGGCACGCGTGGT-3′ |
| GSP2 | 5′-ACTTCCATTGTTCATTCCAC-3′ |
Table II.
Novel λgt10-lacZ-related nucleotide sequences produced from this study
| GenBank® approved accession numbers | Target size (bp) | Description |
| AY927770 | 920 | Spans the mutation in b527 of both the λgt10lacZ and the cloning vector λgt10 |
| AY927771 | 6729 | The λgt10lacZ and λgt10 lom ORF401-like coding sequence (cds) and side fibre genes, and λINT complete cds |
| AY940193 | 6729 | Mus musculus (CD2F1 (BalB/C × DBA/2) transgenic DNA that includes lom and complete cds of side tail fibre genes, and partial cds of the λINT gene |
| AY940194 | 920 | M.musculus genomic DNA template, b527 mutation region |
| DQ101279 | 1011 | M.musculus genomic DNA template rearrangement sequence R1 |
| DQ387054 | 661 | M.musculus genomic DNA template rearrangement sequence R2 |
| DQ387055 | 1097 | M.musculus genomic DNA template rearrangement sequence R3 |
| DQ387056 | 779 | M.musculus genomic DNA template rearrangement sequence R4 |
| DQ387057 | 1001 | M.musculus genomic DNA template rearrangement sequence R5 |
The proposed nucleotide sequence for the λgt10-lacZ transgene model of Muta™Mouse (see Figure 1) is composed of nucleotide sequences considered to be novel by GenBank®, as indicated by new accession numbers above and also nucleotide sequences which have substantial similarity to those previously assigned GenBank® accession numbers λ bacteriophage (J02459), E.coli lacZ (J01636) and imm 434 (M60848; Y00118).
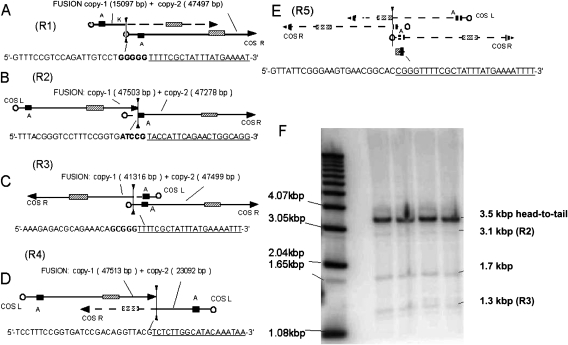
Fig. 2.
Schematic diagrams illustrating in vivo rearrangements of λgt10-lacZ transgene. Panels (A) through (E) show examples of rearranged λgt10-lacZ copies (R1–R5) that correspond to the rearrangement fragments of <1 kb, initially recovered from genomic DNA of three male and two female Muta™Mouse animals by genome scanning methods (see Materials and methods). Donor sequences (underlined) at the fusion site between copies are shown, along with selected features of the transgene: left (COSL) and right (COSR) cohesive ends, genes A, K and lacZ, shown as a hatched box. Additional sequence data can be accessed through GenBank® (see Table II). PCR primers in each case were selected to amplify the fusion fragment and validate these rearrangements in DNA samples from eight males and eight females (see text). Examples R1–R4 were likely formed by head-to-tail (R1 and R2) and head-to-head (R3 and R4) fusions between adjacent transgene copies and stabilisation by sequence loss (dashed lines). Panel E shows example R5 that is composed of 546 bp of lacZ (nucleotides 3371–3916 accession J01636) fused to 113 bp of the bacteriophage lambda A gene (nucleotides 2328–2441 accession J02459) fused to a partial COSL sequence (nucleotides 15–138 accession J02459) and appears to have originated from a fusion and extensive deletion of as many as three transgene copies. Putative crossover sites, shown as bolded sequences in R1–R3, involve similar sequences (e.g. the R1 dimer fusion occurs between sequence GGGGG (nucleotides 15094–15098) and sequence CGGG (nucleotides 15–18), and the latter sequence is also involved in R3 and R5 fusions). (F) A phospho-image of a Southern blot hybridisation derived from agarose gel fractionated EcoRV digested genomic DNA of four animals probed with a 563 bp amplicon spanning the COSL-Nu1 region. DNA ladder fragment sizes are shown on the left of the panel and estimated EcoRV fragment sizes are shown on the right side. In a head-to-tail arrangement of transgenes, the predicted EcoRV fragment would be sized 3.5 kb. The EcoRV fragment sizes that appear to correspond to particular rearranged transgene copies are shown in brackets.
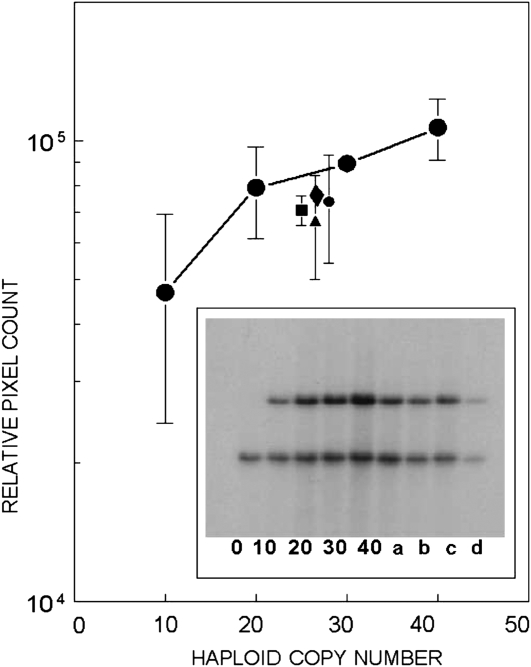
Fig. 3.
Determination of copy number by Southern blot hybridisation. Copy number was determined by quantifying scanned images (pixel counts) of three Southern hybridisation replicates. An example of the primary results is shown in inset figure, which includes standards made up by using a constant amount of CD2F1 non-transgenic mouse DNA alone (0) or mixed with λgt10-lacZ monomer copies ranging from 10 to 40 per haplotype. Male transgenic DNA samples included: bone marrow (a, closed square); heart (b, closed diamond); lung (c, closed circles) and testis (d, closed triangle). Each DNA sample was BamHI digested, electrophoretically fractionated in 0.8% agarose and transferred to nylon membranes before hybridisation in two steps, first to a 32P-labelled λgt10 ori 501 bp probe (upper band) and then to a 32P-labelled 812 bp mouse 18S probe (lower band).
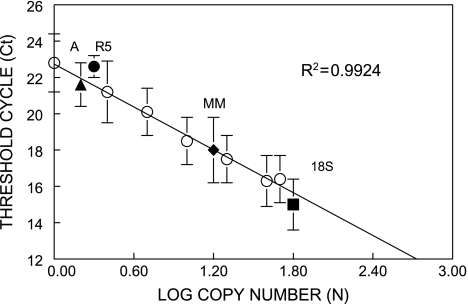
Fig. 4.
Summary of RT–PCR analysis of in vivo transgene copy number. Standard curves were based on quantifications of the ori amplicons, using replicated sets of template composed of CD2F1 non-transgenic mouse DNA mixed with λgt10-lacZ (see Figure 3 Materials and methods) to generate haploid standards of 1, 2.5, 5, 10, 20, 40 and 50 copies. Cycle thresholds (Ct) and efficiency parameters were obtained by iCycler software and show a linear correlation with log copy number values (R2 = 0.9924). Shown are average Ct values for: ori standards (open circles), single-copy annexin V exon 4 (closed triangle, A); R5 rearrangement (closed circle, R5); interpolated copies of Muta™mouse ori (closed diamond, MM) and 18S (closed box, 18S).
Transgene copy determinations by DNA hybridisation
The Southern hybridisation method was performed (19) using 2 μg of Muta™mouse DNA and transgene CD2F1 DNA mixtures (see standards). The DNA was predigested with BamHI endonuclease (10 U at 37°C, 3 h), fractionated by 0.8% agarose gel electrophoresis and transferred to Nytran® SuPerCharge membranes (Schleicher and Schuell, BioScience Inc., Keene, NH, USA) by capillary flow using 0.25 M NaOH and 1.5 M NaCl. Bound DNA was stabilised by ultraviolet cross-linking with 50 mJ per Southern blot (GS Gene Linker, Bio-Rad Laboratories Inc., Hercules, CA, USA). Probes were PCR amplicons of mouse genes and transgene components (see Table I), which were radiophosphate labelled by random priming (20). Hybridisations were carried out with 1 × 106 c.p.m. per ml of hybridisation solution (Ambion, Austin, TX, USA) and washes were carried out at 50°C for 30 min each with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS, with 0.5× SSC-0.1% SDS and at 65°C with 0.1× SSC-0.1% SDS. The amount of probe annealed to each DNA blot was quantified by autoradiography (Kodak Biomax XAR film), followed by scanning and digitalisation of the films to obtain pixel densities (GeneSnap; Syngene Optics Ltd, Cambridge, UK or UN-Scan-it® Ver.5.1 software (Silk Scientific, Orem, UT, USA). Hybridisation results were corrected for probe-specific activity and target size (kilobase pair).
Transgene copy number determinations by RT–PCR
Segments of endogenous genes and transgenes of Muta™Mouse DNA and in CD2F1 DNA-transgene mixtures (see standards) were quantified using the iCycler iQ RT–PCR detection system (Bio-Rad Laboratories Inc.). Oligonucleotide primers (see Table I) were designed with Beacon Designer (3.01 version; Premier Biosoft International, Palo Alto, CA, USA) and synthesised by Cortec DNA Service Laboratories Inc., Kingston, Canada. DNA templates (see previous section) were denatured (100°C, 10 min, followed by 5 min on ice) prior to addition (20 ng) per 20 μl PCR (96-well PCR plates; Bio-Rad Laboratories Inc.). Reactions consisted of 1× iQ™ SYBR® Green Supermix (Bio-Rad Laboratories Inc.): 50 mM KCl, 20 mM Tris–HCl, pH 8.4, 0.2 mM each dNTP, iTaq DNA polymerase, 3 mM MgCl2, SYBR Green (proprietary concentration), 10 nM Fluorescein and 300 nM of specific primers. Amplification was achieved using the following protocol: 40 cycles of 94°C for 15 sec and 58°C for 45 sec. The threshold cycle number (Ct) was calculated with iCycler iQ Optical System Software Ver.3.1 (Bio-Rad Laboratories Inc.).
PCR strategies to obtain in vivo-related transgene fragments
Three strategies were used to scan for putative transgene rearrangements in Muta™Mouse DNA. The first strategy involved the cloning of anomalously sized transgene amplicons that were derived from screening a stock of amplicons generated by PCR. The template was a pool of EcoRV digested Muta™Mouse genomic DNA (∼1.0 to 2.5 kb in size) recovered from agarose gel slices using ZymoClean gel extraction columns (Zymo research Corporation, Orange, CA, USA). The primers spanned key functional regions of the sequenced λgt10-lacZ. The anomalous λgt10-lacZ-related amplicons were identified by agarose gel electrophoresis and further extracted and amplified by inverse PCR (21). After ligation (19), PCRs were carried using COS flanking, antiprimers (see Table I, INV-1 and INV-2) and a proofreading Taq polymerase (LA Taq; Takara Bio Inc., Otsu, Shiga, Japan) to negate potential PCR errors.
The second strategy involved a genome walking procedure (Universal Genome Walker Kit; BD Biosciences, Mississauga, Ontario, Canada). Muta™Mouse DNA was fragmented with different restriction enzymes (EcoRV, DraI, PvuII and SspI) and the resulting fragments were ligated to adaptor nucleotide oligomers (supplied in the kit). Duplicate PCR amplifications were performed with ∼50 ng of each DNA digest, using Advantage genomic polymerase (BD Biosciences), an outer adaptor primer (AP1, provided in the kit), a transgene-specific primer (GW-1 or GW-4 see Table I) and manufacturer’s-specified reaction conditions (7 cycles of 2 sec at 94°C and 3 min at 70°C, followed by 33 cycles of 2 sec at 94°C and 3 min at 65°C and then terminated with 4 min at 67°C). A second amplification (‘nested’ PCR) used 1 μl of a 50× dilution (∼50 ng) of the previous PCR product, an inner adaptor primer (AP2) provided in the kit, a nested transgene-specific primer (GW-2 or GW-3) and manufacturer’s-specified reaction conditions (5 cycles, 2 sec at 94°C, then 3 min at 72°C, followed by 20 cycles, 2 sec at 94°C, then 3 min at 67°C and then terminated with 4 min at 67°C). The PCR products were fractionated by agarose gel electrophoresis, extracted, cloned and sequenced.
The third strategy was modelled after a protocol to identify proviral integration sites (22). PCRs were carried out with SspI linearised Muta®Mouse DNA and a biotinylated λgt10-lacZ-specific primer (BIO-1) and each of the degenerate primers (see FP1–FP4, Table I). Biotinylated PCR products were recovered using streptavidin-coated magnetic beads according to manufacturer’s instructions (Dynabeads kilobase BINDER™ Kit; Invitrogen). A second round of PCRswas performed using 2 μl of purified fragments, the GSP-2 primer (Table I) and a primer representing the 23 nucleotide common region of FP1–FP4 primers. The subsequent PCR products were fractionated by agarose gel electrophoresis, extracted from the gel, cloned and sequenced.
Results and discussion
Status of the λgt10-lacZ sequence in Muta™Mouse
The model for the Muta™Mouse λgt10-lacZ transgene sequence is shown in Figure 1 with details given in the legend. Derivation of the 47 513 bp monomer involved extensive validation of nucleotide sequences of bacteriophage λ, the immunity region of phage 434 (imm 434) and the lacZ-pMC1511 fragment insertion and our novel sequence findings (refer to accession numbers in Table II, Figure 1), which include λgt10 and λNM518 (left arm) and λNM607 (right arm) components (23). Data for these arms clarify ancestral mutations listed for λgt10 genotype (λ srl λ1° b527 srl λ3° imm 434 (srI434+) srl λ4° srl λ5°), the junctions of the lacZ-pMC1511 fragment insertion and imm434.
As a form of validation, key segments of the transgene were amplified, sequenced and compared with corresponding segments from commercial stocks of λgt10 and λCI857 using primer sets mostly listed in Table I. Sequences obtained from separate amplifications of the left arm of λgt10-lacZ and λgt10 (see accession number AY927771) revealed an unreported substitution of open reading frames (ORFs) 401, 314 and 194, with a putative lambdoid Rac prophage tail fibre assembly gene (NCBI GeneID: 946062). The significance of these results is considered in the next section. Further details about the left arm were revealed by analysis of the b527 deletion region. The novel sequence indicates a deletion of 3.6 kb, including ORFs that are not essential for phage viability [e.g. Ea47 (lambdap24), Ea31 (lambdap23), Ea59 and the non-coding att site].
The status of ancestral mutations at the abolished EcoRI sites in λ (accession number J02459, nucleotide positions 21227, 26105, 31748, 39169 and 44973) within ORF314, Ea59, exo, O and S was also clarified by sequencing amplified λgt10-lacZ segments derived from phage plaques (i.e. rescued in vivo copies of the transgene), the commercial λgt10 vector as well as direct amplification of genomic copies from DNA of two transgenic mice. These data also established that the 6.7 kb imm 434 sequence contains regulatory regions N, cI and cro and pMC1511-lacZ sequences, as well as other functionally important regions (e.g. nu1, A, F1, ORF64, S and K). The λgt10 sequences and counterparts of λgt10-lacZ are 98% similar.
Ancestry of the Muta™Mouse transgene
The sequence analysis described in the previous section and Figure 1 provides a new level of detail about this transgene’s ancestry and potential use in Muta™Mouse exposure assays. The finding of substitutions in ORFs 401, 314 and 194, and comparison of them with genes encoding tail fibre proteins of other E.coli lambdoid phage genomes (data not shown), indicate a mosaic pattern consistent with module (gene segment) exchanges between similar genes or clusters of similar genes, which likely originated as a result of illegitimate recombination (24–26). However, these recombination events occurred prior to the λgt10-lacZ construction because the same substitutions were found in the progenitor λgt10 and λCI857. These substitutions would likely have no obvious negative effects on functions relating to the E.coli-based Muta™mouse mutation assay, which includes rescue of monomer copies by in vitro packaging, and infection, replication and virion production necessary for plaque expression.
Potential functional transgene sequence motifs
Databases for murine and E.coli genomes were examined for nucleotide sequence similarities with the full-length transgene model. No similarities with known murine genes were found, but there are numerous CpG islands as well as several putative transcription factor-binding sites distributed along the transgene. At least three or more types of these elements are concentrated within gene A (nucleotide position ∼1300 ± 200 bp) and the 3′ end of J and 5′ end of lom (nucleotide position 17800 ± 200 bp). Earlier observations reported that the transgene is heavily methylated (Cm5) and that the lacZ of this transgene was transcriptionally inactive in Muta™Mouse (1,12). We made similar observations (Shwed, P. unpublished results) and also found two segments that strongly resemble matrix-associated regions (15), in terms of their size and consensus of core sequence. These sequences (see Figure 1, MAR-1 and MAR-2) could potentially serve a role in higher-order chromatin folding, replication and recombination of the transgene and its repeats within the mouse chromosome. Further, the location of MAR-2 is in the vicinity of where rearrangements were found. Given the large size and repetition of the transgene within the mouse genome, the transgene may serve as a model to study agents that cause chromosome disruption and double minutes.
A scan of the transgene revealed that it contains a 5′-GCTGGTGG-3′ motif, called a crossover hot spot instigator (Chi, or χ, reviewed in ref. 27), labelled χ+ at lacZ position 3508 E.coli lacZ (accession J01636) on the antisense strand of the lacZ gene (see Figure 1). This motif would reduce plaque output, but it is antisense with respect to the transgene COS sites and therefore may not be functional (reviewed in ref. 27). The lacZ gene also features three imperfect Chi-motifs, labelled χ0, including one in the sense strand, having the potential to be active by one mutation. In addition, the model 47 513 bp transgene monomer has 71 other χ0 motifs (51 in the forward strand), but not all have been validated. If true, they would further reduce plaque output and plaque-forming unit estimates.
The detailed knowledge of the existing lacZ/E.coli assay presented herein could lead to the development of new assay designs that may result in new insights into the mechanism of mutation induction during E.coli packaging and in the Muta™Mouse genome. Further insights may be gained through detailed studies of the unpublished Muta™Mouse lacZ (E.coli host) mutation database.
Evidence for Muta™mouse transgene rearrangements
Our breeding colony is derived from strain 40.6.13, the pre-commercial progenitor of Muta™Mouse. Molecular characterisation of strain 40.6.13 is not well documented. A Southern blot hybridisation analysis of the founder and F1 progeny indicated an integration of ∼35 copies, possibly at one locus and likely all in head-to-tail configuration (1). The same analysis reported that most copies were likely intact and heavily methylated. An in situ DNA hybridisation analysis of animals from our colony revealed a cluster of transgene copies within the B region of Chromosome 3 (28). However, this method cannot reveal the number or integrity of transgene copies involved.
As an alternate approach towards characterisation of the putative transgene cluster(s), we scanned for presence of aberrant copies using DNA from various tissues of both sexes. In one approach, PCR was used with primers designed to amplify within, and across, sites that would serve as transgene copy junctions, such as COS and MAR (see Figure 1). This analysis revealed minor heterogeneity in the expected size of some PCR targets. In subsequent approaches, the products generated from methods using inverse PCR, genome walking and arbitrary PCR primers were cloned and sequenced using primers described in Table I. From this extensive effort, five types of transgene aberrations were repeatedly detected and sequenced. As summarised in Figure 2A–E and Table II, R1–R4 examples are fusions of at least two copies of the transgene, but R5 is likely a fusion product of as many as three copies. The orientation of transgene copies within R1 and R2 is the same as the bulk of the non-rearranged head-to-tail copies, but R3–R5 are oriented head to head. However, the joining of R3 and R4, through their COS sites (either end) could allow them to be included within a tandem array involving head-to-tail joined R1, R2 or any non-rearranged copy.
The R1–R5 rearrangements were validated in Muta™Mouse DNA by sequencing and use of diagnostic PCR primers and probes relating to the fusion junctions and strand orientation maps (Figure 2A–E). Positive identification of these recombinations in vivo was made for each case by using at least three males and two females and between 6 and 10 DNA samples from three tissues. The example analysis in Figure 2F was derived by using a 614 bp PCR hybridisation probe, which detects both the non-rearranged transgene copies and also R1–R4, due to their size and loss of flanking EcoRV restriction sites near the MAR-2/COS site. The five rearrangements are present in the commercial Muta™mouse colony available from Covance, which indicates that the rearrangements arose early in the lineage, since the Health Canada colony was obtained in 1990.
The transgene copy fusions identified in this study are consistent with complex rearrangements of gene concatemers observed in other transgenic mice (29–32). The fusions could have arisen independently of each other, by either non-homologous end-joining or recombination, as suggested for modifications observed in other types of transgenes after their integration (33,34), or in the case of R2, by sequence degradation due to nuclease ‘nibbling’ (30,35,36). The five types of transgene fusions suggest that a substantial fraction of the transgene array is involved in the rearrangements (at least 10) and these would be defective and not rescued by in vitro packaging, because of size (either <37 kb or >52 kb) and/or loss of key functions (37). No mouse genomic DNA insertion site sequences were recovered as part of this characterisation effort and may reflect a common limitation of these methods to recover very complex or very small transgene-genomic sequence rearrangements. Alternatively, there may be multi-sequence integrity breaks forming one or more transgene loci and chromosomal DNA junctions.
Further studies using DNA from other Muta™Mouse colonies would clarify to what extent these rearrangements are shared and likelihood that they arose during the progenitor construction. Some of these rearrangements can potentially serve as markers within the transgene cluster. As discussed below, R5 is the most reliable one that can be detected by PCR. The integrity scanning methods, along with other strategies [e.g. mapping mutations by restriction site alteration, (11)] could be potentially used to characterise various classes of mutagens such as clastogens and aneugens for effects on transgene copy stability in somatic and germ line cells.
Copy number determinations
The initial report on construction of λgt10-lacZ transgenic founders indicated a copy number range of 3–80 depending on mouse strain, using dot-blot hybridisation methodology, liver samples and a standard composed of λ DNA mixed with tail DNA from an unspecified non-transgenic mouse (1). Strain 40.6 was reported to have 35 copies per haploid genome, while other strains were determined to have different values. However, the Southern blot data suggests that copy number is about the same for all strains. More recently, the copy number for Muta™Mouse has been described as 40 per haploid genome, without reference to how this copy version was derived (38,39). This amount slightly exceeds the averaged estimates by slot blot hybridisation for a cell line (FE1) derived from one of our animals (7). The relatively low transgene copy of FE1 may reflect different measurement methods or that there is actually a copy loss within the FE1. This cell line has been shown to be a mix of karyotypes, having a modal chromosome number of 78, and three transgene loci per cell in 80% of the cells scored (7).
Given the evidence for several defective λgt10-lacZ copies (Figure 2), we determined the haploid copy for animals in our breeding colony using variations in the Southern blot hybridisation method and also RT–PCR. The Southern hybridisation method has been used to monitor founder animals for multiple integration sites and rearrangement by microinjection core facilities (1,10,40). However, based on a large number of experiments, we found that like dot-blot methods, Southern blot methods were not reliable as a quantitative tool when comparing extreme differences in copy number of genes. These analyses included the use of several restriction enzymes to predictably fragment genomic DNA in order to reduce viscosity when titering, fractionating and blotting DNA samples. The example study shown in Figure 3 used BamH1 digested λgt10-lacZ CD2F1 DNA mixtures and DNA of Muta™Mouse tissues probed with both λ ori and 18S ribosomal gene sequences. The relative copy of 18S fragment in both murine genomes was comparable and the ori-associated BamH1 fragments of male and female Muta™Mouse tissues gave copy numbers of 20–30 per haploid genome. However, the analytic conditions for high-copy targets do not reliably detect the single-copy gene. If fused versions of the transgene are in single copy, then based on relative intensities of their hybridisation signals and that of the 3.4 kb non-recombinant hybridised fragment (Figure 2), the number of non-recombinant copies would be ∼25 and the total transgene copies ∼35.
RT–PCR has rapid diagnostic potential and has been used to estimate low-copy transgene insertions in plants (41–43) and more recently, to determine status of transgene zygosity of animals (44–46). To date, few contemporary high-copy eukaryotic transgene copy determinations by RT–PCR have been reported.
To qualify an RT–PCR assay for high-copy estimates, both primer sensitivity and efficiency (as measured by amplicon quantity) were evaluated with specific primer sets described in Table I, using the same reference standards as for the hybridisations, and also annexin V (Exon IV, 675 bp), a single-copy gene that is also located on chromosome 3. As summarised in Figure 4, a reliable standard curve (R2 = 0.9924) was generated with the standards and ori primers that allowed the back-calculation of the copy number for the single-copy annexin gene and copy number of ori amplicons in Muta™Mouse spleen DNA. Additional, RT–PCR comparisons using Muta™Mouse spleen DNA and commercially prepared BigBlue® spleen DNA as templates yielded average copy numbers of 29 ± 4 (n = 3) and 23.5 ± 3.1 (n = 5), respectively. The difference in copy number of the two types of transgenic mice is significant as determined by statistical analysis (Student’s t-test).
The copy number value for BigBlue® spleen DNA is nearly half of the 40 copies previously claimed for BigBlue® mouse (4). However, Barnett et al. (47), who bred Big Blue® mice with non-transgenic mice, suggested that based on mutation frequency of hemizygous offspring with a single marker lacI mutation (41.5 × 10−3), the number of viable shuttle vectors per cell may be as low as 25 (the calculated value was 24.096).
Conclusions
Muta™Mouse has been extensively used over the past 20 years for in vivo mutagenicity assessment [see review by Lambert et al., 2005 (38)], despite the fact that neither the nucleotide sequence nor the integrity of the transgene array, in terms of its tandem multiple copies, were fully characterised. The present study provides a useful molecular-level map of the transgene monomer. This characterisation clarifies previously unknown monomer components, including the nature of ancestral mutations within the λgt10 moiety. Using the monomer model as a baseline and chromosome scanning and cloning methods, at least 10 defective, rearranged copies were found. Of these copies, eight occur as head-to-tail or head to head dimer fusions. One recombinant, R5, probably involves more than two extensively deleted copies and is easily detected by PCR compared to the other rearrangements. Transgene copy determinations by Southern hybridisation and by RT–PCR, indicate that only about two-third of the estimated haploid genome copies can be retrieved and used in E.coli mutation assays. The use of these data and detection methods, as well as the primer list for analysis of external versus endogenous copy standards should enable independent inter-colony comparisons in efforts to establish whether these defects are unique to our colony or the precursor 40.6 strain construction. The R5 could serve as a heterologous single-copy reference marker for future Muta™Mouse studies.
Results of this study further the usefulness of Muta™Mouse to study effects of mutagens at the copy as well as chromosomal structure levels. Further contributions of the study include RT–PCR based methods for detection of transgene gene rearrangements and variable copy number determinations in inter-laboratory comparisons using a standardised reconstructed NCBI mouse genome build, single and multiple copy endogenous genes, and a commercially available reference standard, BigBlue® mouse spleen DNA. The Muta™Mouse transgene construct appears to have potentially utility for assays that detect the effects of clastogens and aneugens, without depending on an E.coli host, for reporting effects.
Funding
Health Canada Genomics R and D; Canadian Regulatory System for Biotechnology funds. Funding for open access charge: Health Canada Genomics R and D; Canadian Regulatory System for Biotechnology funds.
Acknowledgments
We thank K. Nguyen, J. Gingerich and L. Soper for assistance during the initial phase of this project. We thank Drs C. Parfett, G. Pelletier and A. Tayabali for internal review of the manuscript and helpful advice.
Conflict of interest statement: None declared.
References
- 1.Gossen JA, de Leeuw WJ, Tan CH, Zwarthoff EC, Berends F, Lohman PH, Knook DL, Vijg J. Efficient rescue of integrated shuttle vectors from transgenic mice: a model for studying mutations in vivo. Proc. Natl Acad. Sci. USA. 1989;86:7971–7975. doi: 10.1073/pnas.86.20.7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gossen JA, Vijg J. A selective system for lacZ- phage using a galactose-sensitive E. coli host. Biotechniques. 1993;14:330. [PubMed] [Google Scholar]
- 3.Kohler SW, Provost GS, Fieck A, Kretz PL, Bullock WO, Sorge JA, Putman DL, Short JM. Spectra of spontaneous and mutagen-induced mutations in the lacI gene in transgenic mice. Proc. Natl Acad. Sci. USA. 1991;88:7958–7962. doi: 10.1073/pnas.88.18.7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dycaico MJ, Provost GS, Kretz PL, Ransom SL, Moores JC, Short JM. The use of shuttle vectors for mutation analysis in transgenic mice and rats. Mutat. Res. 1994;307:461–478. doi: 10.1016/0027-5107(94)90257-7. [DOI] [PubMed] [Google Scholar]
- 5.Wyborski DL, Malkhosyan S, Moores J, Perucho M, Short JM. Development of a rat cell line containing stably integrated copies of a lambda lacI shuttle vector. Mutat. Res. 1995;334:161–165. doi: 10.1016/0165-1161(95)90007-1. [DOI] [PubMed] [Google Scholar]
- 6.Erexson GL, Cunningham ML, Tindall KR. Cytogenetic characterization of the transgenic Big Blue Rat2 and Big Blue mouse embryonic fibroblast cell lines. Mutagenesis. 1998;13:649–653. doi: 10.1093/mutage/13.6.649. [DOI] [PubMed] [Google Scholar]
- 7.White PA, Douglas GR, Gingerich J, et al. Development and characterization of a stable epithelial cell line from Muta Mouse lung. Environ. Mol. Mutagen. 2003;42:166–184. doi: 10.1002/em.10185. [DOI] [PubMed] [Google Scholar]
- 8.Burkhart JG, Burkhart BA, Sampson KS, Malling HV. ENU-induced mutagenesis at a single A: T base pair in transgenic mice containing phi X174. Mutat. Res. 1993;292:69–81. doi: 10.1016/0165-1161(93)90009-o. [DOI] [PubMed] [Google Scholar]
- 9.Leach EG, Gunther EJ, Yeasky TM, Gibson LH, Yang-Feng TL, Glazer PM. Frequent spontaneous deletions at a shuttle vector locus in transgenic mice. Mutagenesis. 1996;11:49–56. doi: 10.1093/mutage/11.1.49. [DOI] [PubMed] [Google Scholar]
- 10.Nohmi T, Katoh M, Suzuki H, et al. A new transgenic mouse mutagenesis test system using Spi- and 6-thioguanine selections. Environ. Mol. Mutagen. 1996;28:465–470. doi: 10.1002/(SICI)1098-2280(1996)28:4<465::AID-EM24>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 11.Song HL, Jenkins GJ, Ashby J, Tinwell H, Parry JM. The application of the restriction site mutation assay to compare 1-ethyl-1-nitrosourea-induced mutations between the endogenous p53 gene and the transgenic LacZ gene in MutaMouse testes. Mutagenesis. 2001;16:59–64. doi: 10.1093/mutage/16.1.59. [DOI] [PubMed] [Google Scholar]
- 12.Heddle JA, Martus HJ, Douglas GR. Treatment and sampling protocols for transgenic mutation assays. Environ. Mol. Mutagen. 2003;41:1–6. doi: 10.1002/em.10131. [DOI] [PubMed] [Google Scholar]
- 13.Cosentino L, Heddle JA. Effects of extended chronic exposures on endogenous and transgenic loci: implications for low-dose extrapolations. Environ. Mol. Mutagen. 1999;34:208–215. [PubMed] [Google Scholar]
- 14.Ino A, Naito Y, Mizuguchi H, Handa N, Hayakawa T, Kobayashi I. A trial of somatic gene targeting in vivo with an adenovirus vector. Genet. Vaccines Ther. 2005;3:8. doi: 10.1186/1479-0556-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh GB, Kramer JA, Krawetz SA. Mathematical model to predict regions of chromatin attachment to the nuclear matrix. Nucleic Acids Res. 1997;25:1419–1425. doi: 10.1093/nar/25.7.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skopek TR. Of mice and mutants: target size and sensitivity. Mutat. Res. 1995;331:225–228. doi: 10.1016/0027-5107(95)00070-y. [DOI] [PubMed] [Google Scholar]
- 17.Tao KS, Urlando C, Heddle JA. Mutagenicity of methyl methanesulfonate (MMS) in vivo at the Dlb-1 native locus and a lacI transgene. Environ. Mol. Mutagen. 1993;22:293–296. doi: 10.1002/em.2850220419. [DOI] [PubMed] [Google Scholar]
- 18.Taylor AF, Smith GR. Regulation of homologous recombination: Chi inactivates RecBCD enzyme by disassembly of the three subunits. Genes Dev. 1999;13:890–900. doi: 10.1101/gad.13.7.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning a Laboratory Manual. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 20.Feinberg AP, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 21.Ochman H, Gerber AS, Hartl DL. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gentner B, Laufs S, Nagy KZ, Zeller WJ, Fruehauf S. Rapid detection of retroviral vector integration sites in colony-forming human peripheral blood progenitor cells using PCR with arbitrary primers. Gene Ther. 2003;10:789–794. doi: 10.1038/sj.gt.3301935. [DOI] [PubMed] [Google Scholar]
- 23.Huynh TV, Young RA, Davis RW. Constructing and screening cDNA libraries in lambda gt10 and lambda gt11. In: Glover DM, editor. DNA Cloning: A Practical Approach. Oxford, UK: IRL Press; 1985. Vol. 1, pp. 50–51. [Google Scholar]
- 24.Haggard-Ljungquist E, Halling C, Calendar R. DNA sequences of the tail fiber genes of bacteriophage P2: evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. J. Bacteriol. 1992;174:1462–1477. doi: 10.1128/jb.174.5.1462-1477.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Juhala RJ, Ford ME, Duda RL, Youlton A, Hatfull GF, Hendrix RW. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J. Mol. Biol. 2000;299:27–51. doi: 10.1006/jmbi.2000.3729. [DOI] [PubMed] [Google Scholar]
- 26.Montag D, Schwarz H, Henning U. A component of the side tail fiber of Escherichia coli bacteriophage lambda can functionally replace the receptor-recognizing part of a long tail fiber protein of the unrelated bacteriophage T4. J. Bacteriol. 1989;171:4378–4384. doi: 10.1128/jb.171.8.4378-4384.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Myers RS, Stahl FW. Chi and the RecBCD enzyme of Escherichia coli. Annu. Rev. Genet. 1994;28:49–70. doi: 10.1146/annurev.ge.28.120194.000405. [DOI] [PubMed] [Google Scholar]
- 28.Blakey DH, Douglas GR, Huang KC, Winter HJ. Cytogenetic mapping of lambda gt10 lacZ sequences in the transgenic mouse strain 40.6 (Muta Mouse) Mutagenesis. 1995;10:145–148. doi: 10.1093/mutage/10.2.145. [DOI] [PubMed] [Google Scholar]
- 29.Wilkie TM, Palmiter RD. Analysis of the integrant in MyK-103 transgenic mice in which males fail to transmit the integrant. Mol. Cell. Biol. 1987;7:1646–1655. doi: 10.1128/mcb.7.5.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rohan RM, King D, Frels WI. Direct sequencing of PCR-amplified junction fragments from tandemly repeated transgenes. Nucleic Acids Res. 1990;18:6089–6095. doi: 10.1093/nar/18.20.6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki O, Hata T, Takekawa N, Koura M, Takano K, Yamamoto Y, Noguchi Y, Uchio-Yamada K, Matsuda J. Transgene insertion pattern analysis using genomic walking in a transgenic mouse line. Exp. Anim. 2006;55:65–69. doi: 10.1538/expanim.55.65. [DOI] [PubMed] [Google Scholar]
- 32.Kasai F, Yoshihara M, Matsukuma S, O'Brien P, Ferguson-Smith MA. Emergence of complex rearrangements at translocation breakpoints in a transgenic mouse; implications for mechanisms involved in the formation of chromosome rearrangements. Cytogenet. Genome Res. 2007;119:83–90. doi: 10.1159/000109623. [DOI] [PubMed] [Google Scholar]
- 33.Bishop JO. Chromosomal insertion of foreign DNA. Reprod. Nutr. Dev. 1996;36:607–618. [PubMed] [Google Scholar]
- 34.Wurtele H, Little KC, Chartrand P. Illegitimate DNA integration in mammalian cells. Gene Ther. 2003;10:1791–1799. doi: 10.1038/sj.gt.3302074. [DOI] [PubMed] [Google Scholar]
- 35.Hamada T, Sasaki H, Seki R, Sakaki Y. Mechanism of chromosomal integration of transgenes in microinjected mouse eggs: sequence analysis of genome-transgene and transgene-transgene junctions at two loci. Gene. 1993;128:197–202. doi: 10.1016/0378-1119(93)90563-i. [DOI] [PubMed] [Google Scholar]
- 36.Kearns M, Robertson G, Whitelaw E. Sequence analysis of transgene-transgene junctions following microinjection of mouse oocytes. Transgenics. 1995;1:639–647. [Google Scholar]
- 37.Feiss M, Fisher RA, Crayton MA, Egner C. Packaging of the bacteriophage lambda chromosome: effect of chromosome length. Virology. 1977;77:281–293. doi: 10.1016/0042-6822(77)90425-1. [DOI] [PubMed] [Google Scholar]
- 38.Lambert IB, Singer TM, Boucher SE, Douglas GR. Detailed review of transgenic rodent mutation assays. Mutat. Res. 2005;590:1–280. doi: 10.1016/j.mrrev.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Wahnschaffe U, Bitsch A, Kielhorn J, Mangelsdorf I. Mutagenicity testing with transgenic mice. Part I: comparison with the mouse bone marrow micronucleus test. J. Carcinog. 2005;4:3. doi: 10.1186/1477-3163-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reue K, Rehnmark S. Screening transgenic mice by PCR: detection of tandem repeats. Biotechniques. 1994;17:252–253. [PubMed] [Google Scholar]
- 41.Ingham DJ, Beer S, Money S, Hansen G. Quantitative real-time PCR assay for determining transgene copy number in transformed plants. Biotechniques. 2001;31:132–140. doi: 10.2144/01311rr04. [DOI] [PubMed] [Google Scholar]
- 42.Mason G, Provero P, Vaira AM, Accotto GP. Estimating the number of integrations in transformed plants by quantitative real-time PCR. BMC Biotechnol. 2002;2:20. doi: 10.1186/1472-6750-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bubner B, Baldwin IT. Use of real-time PCR for determining copy number and zygosity in transgenic plants. Plant Cell Rep. 2004;23:263–271. doi: 10.1007/s00299-004-0859-y. [DOI] [PubMed] [Google Scholar]
- 44.Tesson L, Heslan JM, Menoret S, Anegon I. Rapid and accurate determination of zygosity in transgenic animals by real-time quantitative PCR. Transgenic Res. 2002;11:43–48. doi: 10.1023/a:1013928600442. [DOI] [PubMed] [Google Scholar]
- 45.Shitara H, Sato A, Hayashi J, Mizushima N, Yonekawa H, Taya C. Simple method of zygosity identification in transgenic mice by real-time quantitative PCR. Transgenic Res. 2004;13:191–194. doi: 10.1023/b:trag.0000026084.32492.eb. [DOI] [PubMed] [Google Scholar]
- 46.Ballester M, Castello A, Ibanez E, Sanchez A, Folch JM. Real-time quantitative PCR-based system for determining transgene copy number in transgenic animals. Biotechniques. 2004;37:610–613. doi: 10.2144/04374ST06. [DOI] [PubMed] [Google Scholar]
- 47.Barnett LB, Tyl RW, Shane BS, Shelby MD, Lewis SE. Transmission of mutations in the lacI transgene to the offspring of ENU-treated Big Blue male mice. Environ. Mol. Mutagen. 2002;40:251–257. doi: 10.1002/em.10114. [DOI] [PubMed] [Google Scholar]