Figure 6.
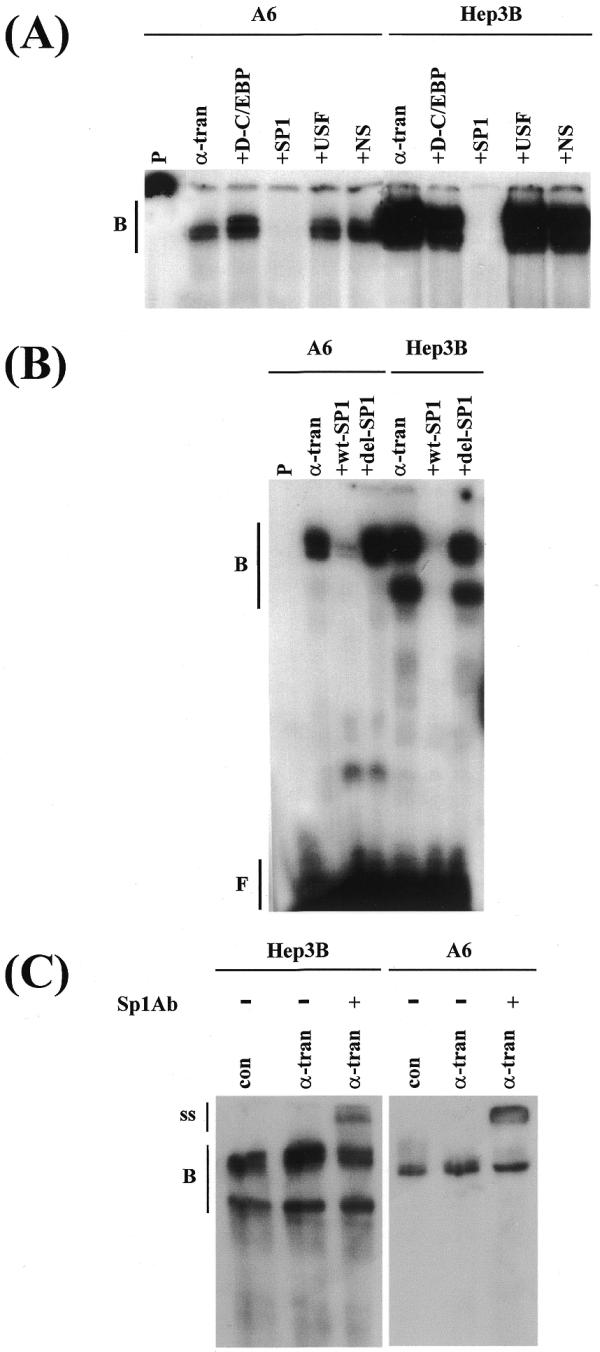
Binding of Sp1 to the –82/+41 region. EMSA were carried out using nuclear extracts from Hep3B or A6 cells that had been transfected with the pCS2+ control vector (con) or the pCS2+xα expression plasmid (α-tran) and either –82/+41 promoter fragment (A and B) or an oligonucleotide containing the putative Sp1 recognition sequence (C). Competition experiments used a 500-fold excess of oligonucleotides for: a high affinity C/EBP binding site (+D-C/EBP), consensus Sp1 recognition sequence (+SP1), consensus USF binding site (+USF), sequences corresponding to the –53 to –81 region of the xC/EBPα promoter (+NS), the Sp1 site from the xC/EBPα promoter (+wt-SP1) or a larger sequence containing deletion of this site (+del-SP1). In (C), + indicates the profile obtained with extracts from pCS2+xα-transfected cells in the presence of antiserum against Sp1. Vertical lines labelled B, ss and F show the position of the DNA–protein complex, the antibody–DNA–protein-supershifted complex and the free probe, respectively.
