Lesion-specific DNA glycosylases play the key role in base excision DNA repair of finding the damaged base and catalyzing its removal1–3. Structural4–6, computational6,7 and experimental studies8,9 have shown that when MutM, a bacterial DNA glycosylase specific for 8-oxoguanine (oxoG), encounters an oxoG lesion, the damaged nucleobase is extruded from the DNA and inserted into the enzyme active site, whereupon catalysis of glycosidic bond cleavage ensues. Potential of mean force (pmf) calculations6 making use of X-ray crystal structures of MutM bound to DNA containing either an extrahelical oxoG bound in the enzyme active site4,5, or an intrahelical oxoG6 or G4, have shown that the free energy barrier for extrusion of oxoG is lower by 7 kcal/mol than that for G. To obtain a crystal of a “lesion-recognition” complex (LRC) with the oxoG in the active site of MutM bound to DNA5, the catalytically essential Glu3 (E3) residue was mutated to Gln (Q) (MutME3Q)10 (Fig. 1). In the pmf simulations, the Q3 residue was restored computationally to E3 to obtain the wild-type enzyme (MutMWT). The simulations indicated that extrusion of the oxoG into the active site was disfavored by ~5 kcal/mol compared to the intrahelical state. Why then had it been possible to crystallize such an apparently disfavored LRC? Here, we show that the E3Q mutation provides substantial stabilization of oxoG in the active site of MutM. The simulations show further that oxoG is stabilized in the active site by protonation of the E3 carboxyl group during the insertion process. It is pointed out also that the protonation could be significant for base-excision catalysis. The results highlight the importance of accounting for the potential energetic and structural effects of mutations frequently introduced into enzymes to capture otherwise fleeting intermediates11,12.
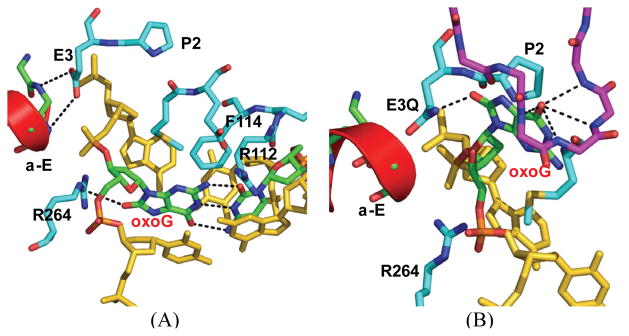
Figure 1.
Comparison of the MutM structures of (A) the encounter complex (EC) and (B) the lesion recognition complex (LRC). The oxoG-capping loop in LRC is shown in purple sticks, and the α-E helix is shown in red cartoon. In the base extrusion process, oxoG is rotated along its glycosidic bond to the syn conformation.
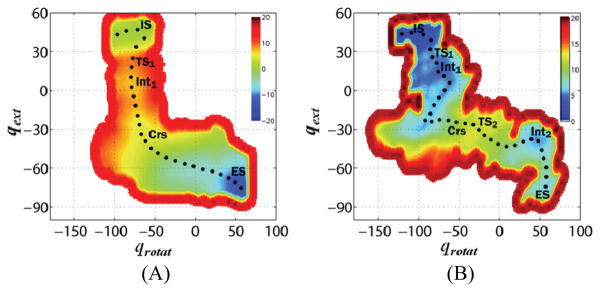
Pmf simulations13 for the extrusion of oxoG by MutME3Q were performed, using the method described previously6 for MutMWT (see the Supporting Information). Figures 2 and S1 show the comparison of the pmf profile for MutME3Q with that of MutMWT having E3 deprotonated (denoted as E3-COO−). The free energy for oxoG extrusion into the active site of MutME3Q is reduced by a significant amount (−14.7 kcal/mol), relative to that for MutMWT, so that oxoG is more stable in the active site of the E3Q mutant than in the intrahelical configuration, in agreement with experiment. The shape of the two-dimensional pmf is also different for the wild-type and the E3Q mutant (Fig. 2). For MutMWT, the rate-limiting barfrier (11 fkcal/mol at TS2) is for the anti to syn rotation of the glycosidic bond, while for MutME3Q, the rotation after extrusion occurs without a barrier and the free energy decreases continuously until oxoG is in the enzyme active site. The barrier for the first step (TS1), corresponding to the extrusion of the base through the minor groove, is 4.2 kcal/mol for MutMWT and 8.7 kcal/mol for MutME3Q. Pmf simulations for G extrusion show that the E3Q mutation also reduces the free energy of G binding to the active site to −12.1 kcal/mol, relative to that of the intrahelical conformation.
Figure 2.
Computed potentials of mean force for oxoG extrusion, rotation, and entrance into the active site of (A) MutME3Q and (B) E3-COO− MutMWT. The pseudo-dihedral angles for the base extrusion and rotation are qext and qrotat, respectively, which represent the extrusion of oxoG into active site from the intrahelical state (IS) to the active site bound state (ES); for the definition of the pseudo-dihedral angles, see Fig. S2. Along the minimum free energy path (shown by black dots), several key points are indicated: transition states (TS1 for the base extrusion from the DNA helix and TS2 for the base rotation), intermediate states (Int1 for the extrahelical base and Int2 for the state where oxoG forms a hydrogen bond with P2)6, and the crossover point (Crs) (see Fig. 3). The unit for dihedral angles is degrees and for free energy is kcal/mol.
Free energy simulations using λ-dynamics-based thermodynamic integration (TI)14,15 were carried out to check the pmf results. The TI values confirm that the E3Q mutation significantly lowers the free energies for both oxoG and G binding in the enzyme active site (see Table S1). Free energy components analysis16 (Fig. S3) further shows that the dominant destabilization arises from the deprotonated E3 in the active site, which interacts repulsively with the extruded oxoG and neighboring phosphate groups. These interactions are less repulsive for Q3, as expected.
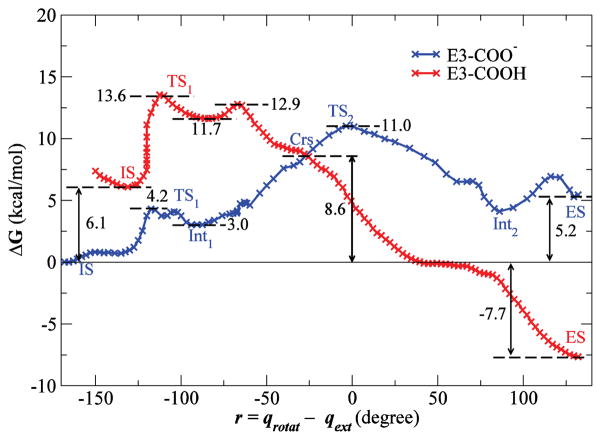
The above results raise the question why oxoG is favored in the active site of the wild-type enzyme. One possibility is that E3 becomes protonated when oxoG enters the active site, although it has been assumed that this residue is deprotonated at pH 7. The crystal structure suggests that the repulsive interactions of E3-COO− with phosphate groups of DNA5 could be diminished by neutralization of the negative charge on E3. Further, a hydrogen bond between the protonated E3 (denoted as E3-COOH) and the 8-oxocarbonyl of the extrahelical oxoG can be formed in the active site; this is analogous to the hydrogen bond formed between 8C=O and the side-chain NH2 group of Q3. The free energy profiles for extrusion of oxoG for E3-COO− and E3-COOH are shown in Fig. 3 (the corresponding results for G are shown in Fig. S4). The two pmf profiles cross at an early stage of the base rotation; the progress variable r defined in the figure is equal to −26° for oxoG and −24° for G at the crossover point (denoted as Crs in Figs. 3 and S4). Since the system can cross from one free energy surface to the other by proton transfer, the free energy for oxoG extrusion into the active site is −7.7 kcal/mol (with E3-COOH), and the rate-limiting barrier is at Crs (8.6 kcal/mol). Protonation of the E3 residue also stabilizes G in the active site, but G is still more stable in the intrahelical state (0.6 kcal/mol) and the free energy barrier for its extrusion is 11.0 kcal/mol at Crs. Alchemical TI simulations for the protonation of E3 are consistent with the results from pmf simulations (Table S2).
Figure 3.
Potentials of mean force along a progress variable (r) equal to the difference qrotat and qext for oxoG extrusion by MutMWT with a protonated (red) or deprotonated (blue) E3 residue. The profiles for E3-COOH are shifted upward by 6.1 kcal/mol, the free energy required to protonate E3 residue at pH 7 (see Supporting Information). Indicated points correspond to points labeled in Fig. 2 and Fig. S5. The zero of free energy is that of the intrahelical state (IS).
In the search of the DNA for the damaged base by MutM, a fast oxoG recognition mechanism is required because of the high speed of MutM sliding along DNA9. Previous results6 have shown that the initial extrusion of the base through the minor groove of the DNA helix is indeed fast and discriminates oxoG from G kinetically. The present results suggest that there also could be a thermodynamic discrimination step in the active site. If E3 becomes protonated, the free energy for binding of the extruded oxoG in the active site is favorable (−7.7 kcal/mol, Fig. 3), while it remains unfavorable for G (0.6 kcal/mol, Fig. S4). The calculated effects correspond to a difference of a factor of 108 in the equilibrium constant between oxoG and G in the active site. The results suggest the following sequence of events for extrusion of oxoG into the active site: (1) E3 is initially deprotonated when oxoG is extruded from the DNA (TS1 barrier of 4.2 kcal/mol); (2) once the system reaches an intermediate extrahelical state (Int1, 3.0 kcal/mol), oxoG begins rotating around its glycosidic bond and E3 becomes protonated (Crs, at the crossover point of 8.6 kcal/mol); and (3) oxoG enters the active site without any additional barriers. There are a number of waters in the protein-DNA interface4,6 that could serve as the source of the proton; also, Pro2, which has to be deprotonated to be a nucleophile, could provide the proton through a water chain near the interface17.
In the present study, we have investigated the effect of the alterations of the key residue E3 in MutM on the free energy along the nucleobase extrusion pathway. We conclude that the E3Q mutation favors oxoG binding in the enzyme active site compared to the wild-type enzyme. Analyses of the computed free energies for MutMWT provide new insights into the role of the E3 residue in base extrusion and recognition of a damaged-nucleobase. The similarity between the side chains of Q and protonated E and the pmf simulations suggest that MutM exploits a two-step discrimination mechanism: a fast kinetic discrimination at the early step of the damaged-base recognition and a thermodynamic discrimination by protonating the E3 residue, which favors binding of oxoG in the active site. The glycosyl transfer reaction has been suggested to occur via a SN1-like displacement of oxoG by the nucleophile Pro25; if protonated, E3 would facilitate the reaction by activating the oxoG leaving group, for example, through the protonation at O8, and subsequently by stabilization of the oxocarbenium ion. Further experimental and computational studies are required to confirm the present results concerning lesion recognition and catalysis by this important enzyme.
Supplementary Material
Acknowledgments
This work is supported by grants from NIH to G.L.V. (CA100742) and M.K. (GM030804), and by a postdoctoral fellowship from National Cancer Center to K.N. NERSC and FAS at Harvard University provided computing resources. K.N. acknowledges Drs. J. Pu, V. Ovchinnikov, G. Zheng, and M. Spong, and Ms. Y. Qi for helpful discussion.
Footnotes
Supporting Information Available: Computational details and table and figures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Lindahl T, Wood RD. Science. 1999;286:1897. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 2.Krokan HE, Nilsen H, Skorpen F, Otterlei M, Slupphaug G. FEBS Letters. 2000;476:73. doi: 10.1016/s0014-5793(00)01674-4. [DOI] [PubMed] [Google Scholar]
- 3.Fromme JC, Banerjee A, Verdine GL. Curr Opin Struct Biol. 2004;14:43. doi: 10.1016/j.sbi.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Banerjee A, Santos WL, Verdine GL. Science. 2006;311:1153. doi: 10.1126/science.1120288. [DOI] [PubMed] [Google Scholar]
- 5.Fromme JC, Verdine GL. J Biol Chem. 2003;278:51543. doi: 10.1074/jbc.M307768200. [DOI] [PubMed] [Google Scholar]
- 6.Qi Y, Spong M, Nam K, Banerjee A, Jiralerspong S, Karplus M, Verdine GL. 2009 doi: 10.1038/nature08561. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song K, Kelso C, de los Santos C, Grollman AP, Simmerling C. J Am Chem Soc. 2007;129:14536. doi: 10.1021/ja075128w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minetti CASA, Remeta DP, Breslauer KJ. Proc Natl Acad Sci USA. 2008;105:70. doi: 10.1073/pnas.0710363105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS. Proc Natl Acad Sci USA. 2006;103:5752. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lavrukhin OV, Lloyd RS. Biochemistry. 2000;39:15266. doi: 10.1021/bi001587x. [DOI] [PubMed] [Google Scholar]
- 11.Fersht AR, Winter G. Ciba F Symp. 1985;111:204. doi: 10.1002/9780470720929.ch14. [DOI] [PubMed] [Google Scholar]
- 12.Fersht AR. Structure and mechanism in protein science: a guide to enzyme catalysis and protein folding. W. H. Freeman; New York: 1999. [Google Scholar]
- 13.Torrie GM, Valleau JP. J Comput Phys. 1977;23:187. [Google Scholar]
- 14.Kong X, Brooks CL., III J Chem Phys. 1996;105:2414. [Google Scholar]
- 15.Kollman PA. Chem Rev. 1993;93:2395. [Google Scholar]
- 16.Boresch S, Karplus M. J Mol Biol. 1995;254:801. doi: 10.1006/jmbi.1995.0656. [DOI] [PubMed] [Google Scholar]
- 17.Cui Q, Karplus M. J Phys Chem B. 2003;107:1071. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.