Figure 1. NhaA - NHAoc/NHA2 sequence alignment.
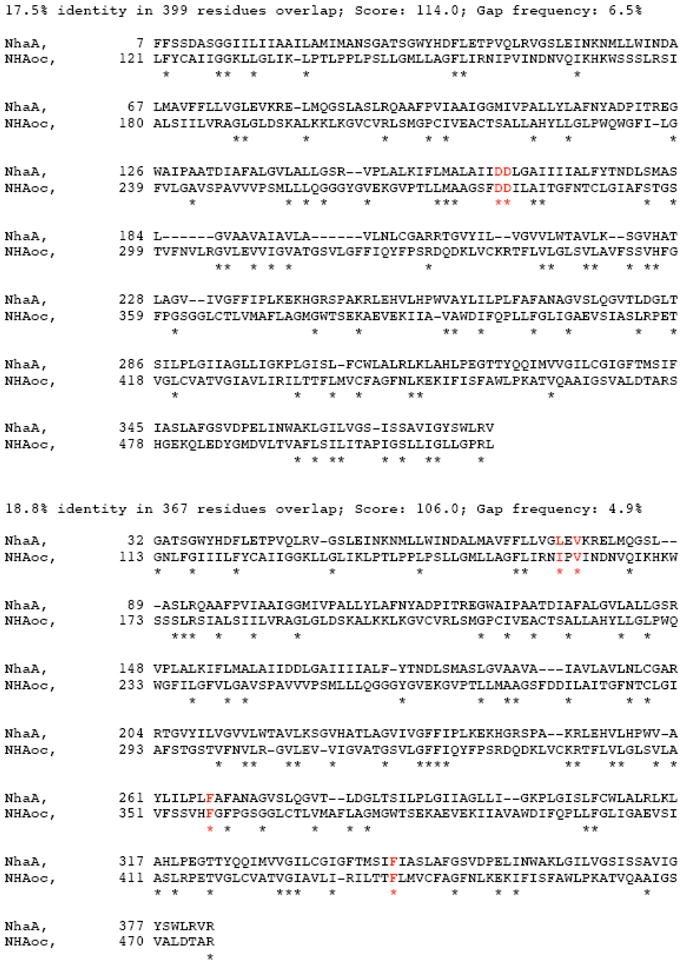
The alignments are reported in order of decreasing similarity score and share no aligned pairs. We aligned NhaA and NHAoc/NHA2 using the BLOSUM30 comparison matrix with the following parameters: Number of alignments to be computed: 20; Gap open penalty:12, Gap extension penalty:4. Above are two of the returned alignments indicating with a “*” the conserved amino acids. Also highlighted (in red): NHAoc/NHA2 Asp278-Asp279, I159, V161, F357 and F437 as well as their matching NhaA residues. Asp278-Asp279 are homologous to Asp163 - Asp164 of NhaA, which are essential for antiport activity. I159, V161 and F357 are amino acids that are substituted as a result of human SNPs. F357 is homologous to F267 of NhaA and amino acid that regulates the stoichiometry of NhaA.
