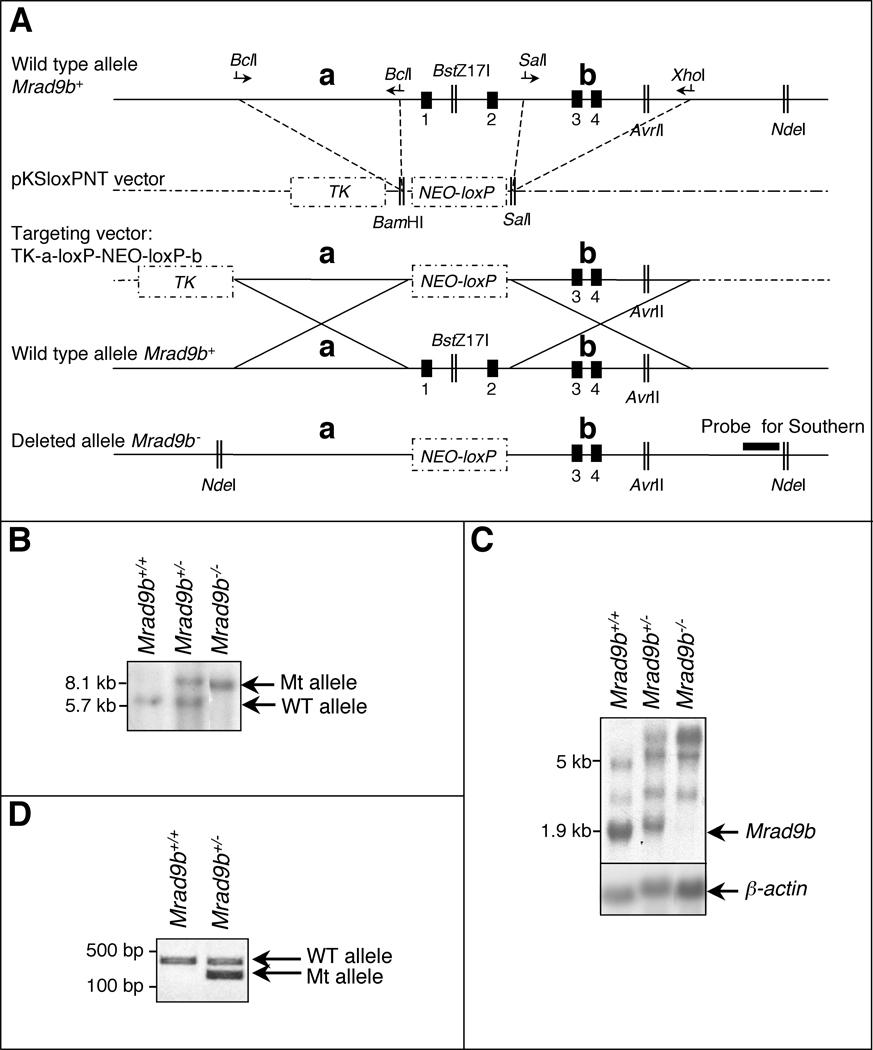
Figure 1.
(A) Targeted disruption of the mouse Mrad9b gene. Vector-derived sequences are shown as stippled lines; genomic DNA illustrated as continuous lines. Arrows represent primers for PCR amplification; Vertical black boxes (1–4) are the first four exons of Mrad9b. NEO, neomycin resistance gene. TK, thymidine kinase. a and b are two Mrad9b genomic fragments that were amplified by PCR, ligated into pKSloxPNT vector and used for homologous recombination between the targeting construct and the corresponding genomic region. Areas of homology are depicted by large “Xs”. The targeting vector was linearized with AvrII before transfection into ES cells. (B) Southern blot analysis of Mrad9b in Mrad9b+/+, Mrad9b+/− (clone #341) and Mrad9b−/− mouse ES cells. The pattern for Mrad9b+/− (clone #350) was the same as for clone #341 (data not shown). Genomic DNA was digested with NdeI and BstZ17I, separated on an agarose gel, and probed. (C) Northern blot analysis of Mrad9b expression in Mrad9b+/+Mrad9b+/− (clone #341) and Mrad9b−/− mouse ES cells. Expression in Mrad9b+/− ES cell clone #350 (data not shown) was similar to that in clone #341. Beta–actin served as the internal control. (D) Genotyping of mice by PCR using tail DNA. WT, wild type; Mt, deletion mutant.