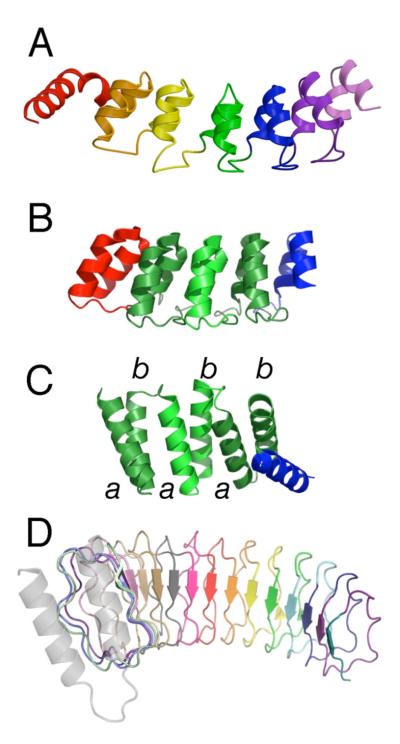
Figure 1. The modular architectures of repeat proteins.
(A) Crystal structure of the Notch ankyrin domain (1ot8.pdb, chain A) consisting of six structured ANK repeats (sequence repeats 2-7) and an N-terminal partly structured repeat. (B) Crystal structure of a consensus ankyrin repeat protein (2qyj.pdb) containing three consensus repeats (green) and N- and C-terminal caps (red and blue respectively). (C) Crystal structure of a consensus-based TPR protein (1na0.pdb, chain A) containing three consensus repeats and a C-terminal cap (blue). (D) Crystal structure of YopM, a leucine-rich repeat protein containing 15 full LRR repeats of the bacterial subtype (15jl.pdb). For naturally occurring (heterogeneous) proteins, individual repeats are shown in different colors; selection of boundaries between repeats (color changes) is somewhat arbitrary, and is based on considerations such as intron position, interresidue contact density, surface area, and visual impression. For the consensus ankyrin and TPR proteins, consensus repeats are shown with the same color but alternate in color saturation. This figure was prepared using PyMol (DeLano, 2003).