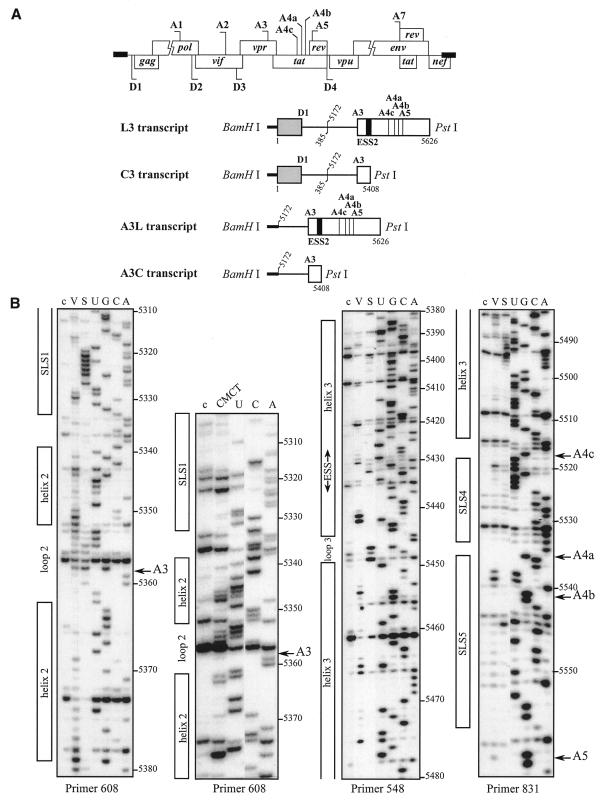
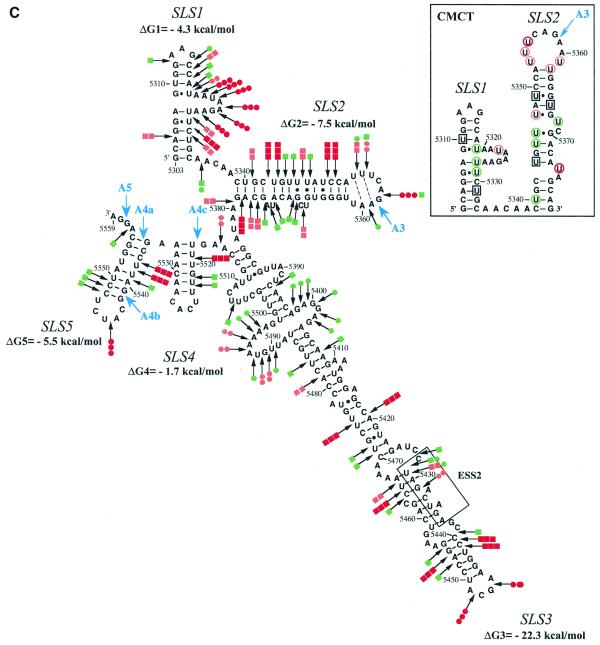
Figure 1.
Secondary structure analysis of the HIV-1/BRU RNA region from positions 5303 to 5559. (A) Schematic representation of the HIV-1 genome and of the RNAs used for secondary structure analysis. In the HIV-1 genome, the 5′ss (D) and 3′ss (A) are shown; boxes, open reading frames. In the four RNA transcripts (L3 to A3C), numbering of HIV-1/BRU RNA sequences is according to Ratner et al. (25), exon sequences are represented by rectangles, introns by thin lines, the thick horizontal lines at the beginning of the transcripts correspond to sequences generated by plasmid pBluescriptKSII+. The 5′ss, 3′ss and ESS2 are indicated. In transcripts C3 and L3, the junction between the two HIV-1 RNA regions within the intron is indicated by a vertical broken line. (B) Examples of primer extension analyses of enzymatically digested and chemically modified L3 transcript. Lanes marked by V, S and CMCT correspond to V1 RNase digestion, S1 nuclease digestion and CMCT modification, respectively. Conditions for digestion and modification are given in Materials and Methods. Lanes marked by c correspond to control experiments with the untreated RNA transcript, lanes UGCA to the sequencing ladders. Numbering of nucleotides in the HIV-1/BRU RNA is on the right of the autoradiogram, as well as the positions of the 3′ss (A3, A4c, A4a, A4b and A5). Positions of the various stem–loop structures identified on the basis of this analysis (Fig. 1C) and of ESS2 are shown on the left. The primers used for extension with reverse transcriptase are indicated below each autoradiogram. (C) The results of enzymatic probing are schematically represented on the proposed secondary structure model. Cleavages by enzymes are shown by arrows surmounted with circles for S1 nuclease and squares for V1 RNase. Three red circles or squares indicate a strong cleavage, two orange circles or squares a medium cleavage, one green circle or square a low cleavage. The free energy of the proposed stem–loop structures at 37°C, in 1 M NaCl were calculated with the MFold software. Positions of nucleotides in the HIV-1/BRU RNA are given. Stem–loop structures are designated as SLS1–5. The various 3′ss are indicated by blue arrows. The ESS2 inhibitory element is squared. Results of CMCT modification of SLS1 and SLS2 are shown in the inset. Red circles indicate a strong, orange a medium and green a low level of modification. Squared nucleotides were not modified.