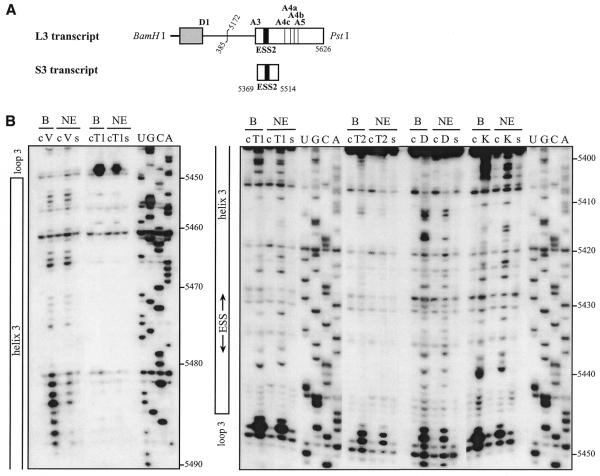
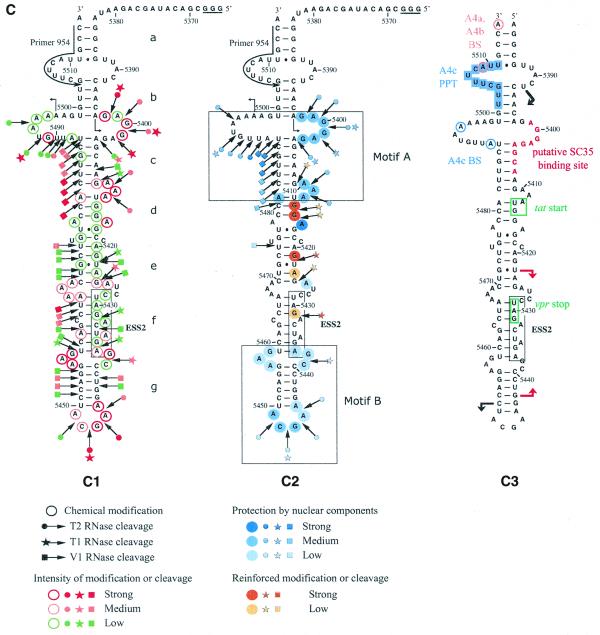
Figure 4.
The HIV-1/BRU SLS3 is formed in a nuclear extract and two structural motifs A and B are protected by association with nuclear components. (A) The S3 transcript used for the experimental analysis is shown. (B) Examples of primer extension analyses of the S3 transcript cleaved by V1, T1 or T2 nucleases (lanes marked by V, T1, T2, respectively) or modified by kethoxal or DMS (lanes marked by K or D, respectively) are presented. Enzymatic and chemical reactions were either performed in the splicing buffer D (B) or in a nuclear extract (NE). As a control, a primer extension was made with the intact RNA transcript incubated in the absence of reagent, either in buffer D or in the nuclear extract (lanes marked by c). Lanes UGCA correspond to the sequencing ladders. Numbering of the nucleotides in the HIV-1/BRU RNA is indicated on the right. Positions of the loops and helices 3 and of ESS2 are shown on the left. (C) Schematic representation of results of chemical and enzymatic probing of S3, in the splicing buffer D (C1) or in the nuclear extract (C2). In Panel C1, the circled nucleotides were modified by DMS or kethoxal. Cleavages by RNases are shown by arrows, surmounted with a circle for T2 RNase, a star for T1 RNase and a square for V1 RNase. Green, orange and red symbols indicate low, medium and strong modifications or cleavages, respectively. In C2, protection against the action of chemical reagents and nucleases are shown in blue, the intensity of the blue color reflects the level of protection. Increased sensitivity to chemical reagents and nucleases is indicated in yellow (low increase) or orange (medium increase). In both C1 and C2, the oligonucleotide primer 954 used for the reverse transcriptase analysis is indicated. The 3G residues at the 5′-end of the S3 transcript were generated by the T7 RNA polymerase promoter. The portion of the S3 RNA that was analyzed is delimited by the two broken arrows in C1. Due to pause of reverse transcriptase at some of the V1 cleavage sites in the region from positions 5397 to 5438 in the nuclear extract, no estimation of their variation of intensity as compared to naked RNA is given for this part of SLS3. In C3, the HIV-1/BRU functional sequences contained in SLS3 are indicated, namely: the tat start codon and vpr stop codon (squared in green), the putative SC35 binding site (red letters), the branched sites (circled in blue) and PPT (in blue rectangles) of the A4c 3′ss and the branched sites for A4a and b 3′ss (circled in orange). The limits of the RNA fragments used by Caputi et al. (11) and Del Gatto-Konczak et al. (32) for hnRNP A/B crosslinking experiments are indicated by black and red arrows, respectively.