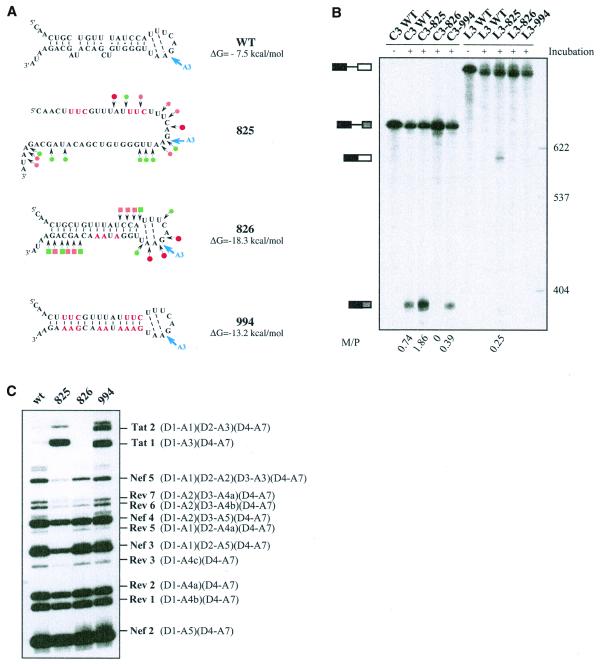
Figure 5.
Effects of alteration of SLS2 on site A3 efficiency in vitro and in vivo. (A) The generated mutations (red nucleotides) and the expected structure of the variant RNAs are shown. For variants C3.825 and C3.826, the secondary structure was probed experimentally, results are schematically represented using the same symbols as in Figure 2C. The free energies of the structures at 37°C in 1 M NaCl, as calculated by the MFold program, are given. (B) PAGE of the in vitro splicing products of the wild-type and variant C3 and L3 transcripts. Uniformly labeled RNAs, prepared as described in Materials and Methods, were incubated for 150 min in a HeLa cell nuclear extract in splicing conditions. Untreated C3 and L3 RNAs were fractionated in parallel, as well as size markers (molecular weights on the right of the panel). Positions of the C3 and L3 transcripts and of their spliced products are indicated on the left of the panel. The M:P ratios (given below the lanes) were calculated by estimation with a phosphorimager of the radioactivity in the bands of gel corresponding to the mature RNA and the residual transcript, respectively. (C) PAGE of the RT–PCR products obtained by amplification of total RNA extracted from HeLa cells transfected with the wild-type or the mutated mini-proviral cDNA pΔPSP, with the oligonucleotide primers BSS and SJ4.7A (see Materials and Methods). Nomenclature of the RT–PCR products is according to Purcell and Martin (1).