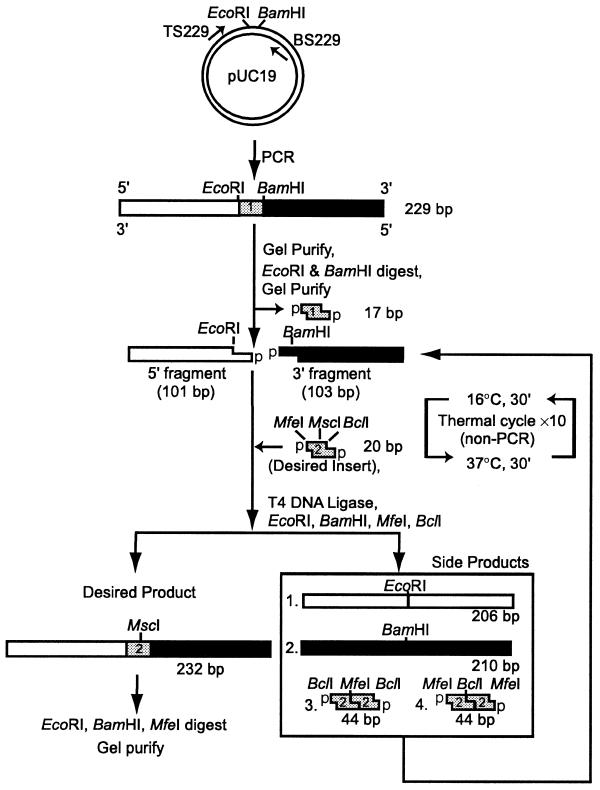
Figure 1.
Strategy used to construct DNA molecules. A 229 bp PCR fragment that contains EcoRI and BamHI cleavage sites in the middle was generated from pUC19 by 20 rounds of PCR. Primer sequences are given in Materials and Methods. After PCR the sample was purified on an agarose gel (Zymoclean) and digested with EcoRI and BamHI to release a 17 bp fragment (insert 1, gray). The 5′-fragment (101 bp) containing an EcoRI sticky end (white) and 3′-fragment (103 bp) containing a BamHI sticky end (black) were gel purified. The desired insert (insert 2, gray) with a MfeI site on one end and a BclI site on the other was inserted between the 5′- and 3′-fragments using T4 DNA ligase. Inserts used in these experiments contained unmodified dsDNA, single bulged A-containing dsDNA or ribose-containing dsDNA. All inserts were 20 bp and had four 5′-dangling nucleotides on each end with a 5′-phosphate, denoted with a p. Since the desired product with a single insert has lost all four restriction sites, it can be enriched from the four side-products by digestion with the four restriction enzymes, followed by agarose gel purification (Zymoclean). Ten rounds of thermal cycling were used to recycle the side-products and increase the overall yield.