Figure 1. I-CreI-Generated Double-Strand Breaks Create Chromosome X Fragments that Lag at Anaphase.
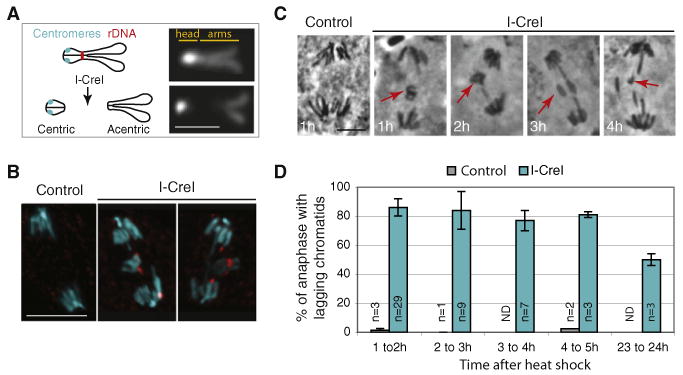
(A) Schematic illustration (left) and DAPI staining (right) of the Drosophila mitotic X chromosome. The centromeres (cyan circles) are very close to the telomeres in the heterochromatin “head” brightly stained with DAPI. The endonuclease I-CreI cuts at the rDNA locus located close to the centromeric heterochromatin. I-CreI generates two distinct chromosome fragments, a small heterochromatic fragment containing the centromeres (centric) and a long fragment containing the sister chromatid arms (acentric). The scale bar represents 2 μm.
(B) DAPI (cyan) and anti-γH2Av (red) fluorescent images of anaphase neuroblasts from heat-shocked third-instar larvae, with (I-CreI) or without (control) the I-CreI transgene. γH2Av that marks DSBs is detected on the lagging chromatids upon I-CreI induction. The images are maximum projections of deconvolved Z sections. The scale bar represents 10 μm.
(C) Orcein staining of anaphase neuroblasts from heat-shocked third-instar larvae with (I-CreI) or without (control) the I-CreI transgene. The time the larvae were dissected after heat shock is indicated on the bottom left. Lagging chromatids (red arrows) are observed after I-CreI induction. The scale bar represents 5 μm.
(D) Frequency of anaphase neuroblasts described in panel C with lagging chromatids (mean ± STD, n = number of brains).
