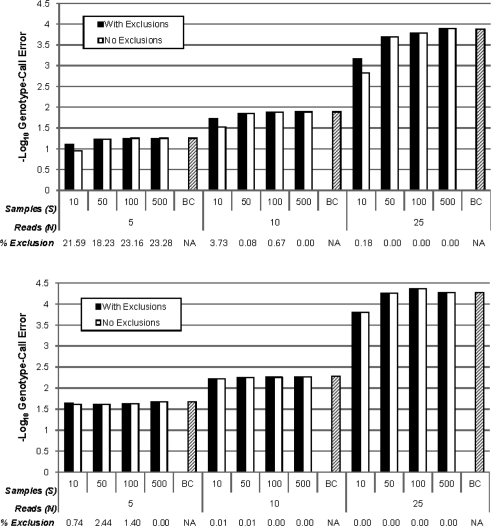
Fig. 4.
Simulated genotype-call error rates (− log 10-scaled) for SeqEM assuming HWE in populations in HWE. Higher values on the − log 10 scale correspond to lower error rates. We considered a nucleotide-read error rate of α = 0.01, sample sizes of S = 10, 50, 100 and 500, read depths of N = 5, 10 and 25 and allele frequencies of p = 0.5 (top) and 0.05 (bottom). Results are presented for all variants as well as excluding variants flagged as potentially poorly modeled by our heuristic (no EM convergence within 100 iterations or nucleotide-read error rate estimate exceeding 0.1). Percentage exclusion indicates the percentage of flagged variants. Expected genotype-call error rates for the Bayes classifier (BC) using true parameter values (Fig. 3) are also included as a sample size of BC.