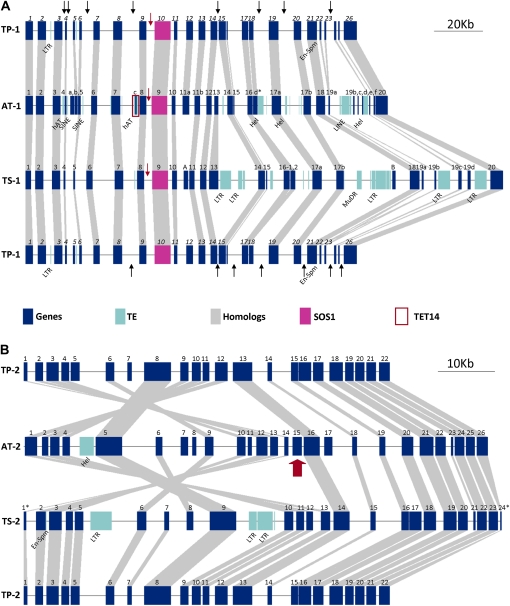
Figure 2.
Genome organization for TP-1 (A) and TP-2 (B) in comparison with the organization of orthologous regions in Arabidopsis and T. salsuginea. A, Black arrows in TP-1 show deletions in comparison with AT-1 or TS-1. Red arrows indicate the intergenic space between SOS1 orthologs and their nearest 5′ gene model. B, The inversion break point is indicated by a red arrow. Symbols for the genes are as in Supplemental Tables S1 and S2. Symbols used for transposable elements (TEs) are LTR (retrotransposon with long terminal repeats), SINE (short interspersed transposable elements), LINE (long interspersed transposable elements), MuDR (Mutator-like DNA transposon), Hel (helitron), En-Spm (Enhancer-Suppressor/Mutator transposon), and hAT (hAT family DNA transposon).