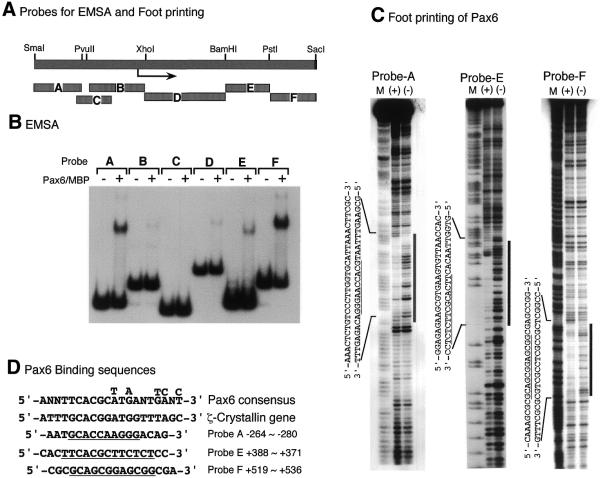
Figure 6.
Identification of Pax6-binding sites of the c-maf gene. (A) Schematic representation of the probes used for EMSA and footprinting analyses. (B) EMSA of c-maf promoter fragments with Pax6–MBP fusion protein. The terminally labeled fragments indicated were bound with Pax6–MBP protein (100 ng) and analyzed as described in Materials and Methods. (C) Footprinting analysis of c-maf promoter fragments. Probe A, E or F was mixed with Pax6–MBP protein (1 µg), treated with DNase I and analyzed by denatured polyacrylamide gel electrophoresis. The same probes, modified and digested by the Maxam–Gilbert method, were used as makers (M). (+) and (–) indicate with or without Pax6–MBP protein, respectively. Footprinting regions are indicated by vertical bars and lined under the sequences. (D) Pax6-binding sequences. Pax6 paired domain binding consensus sequence obtained by PCR-based oligonucleotide selection (33) and that of the promoter region of the ζ-crystallin gene (28) are aligned together with binding sequences of the c-maf promoter.