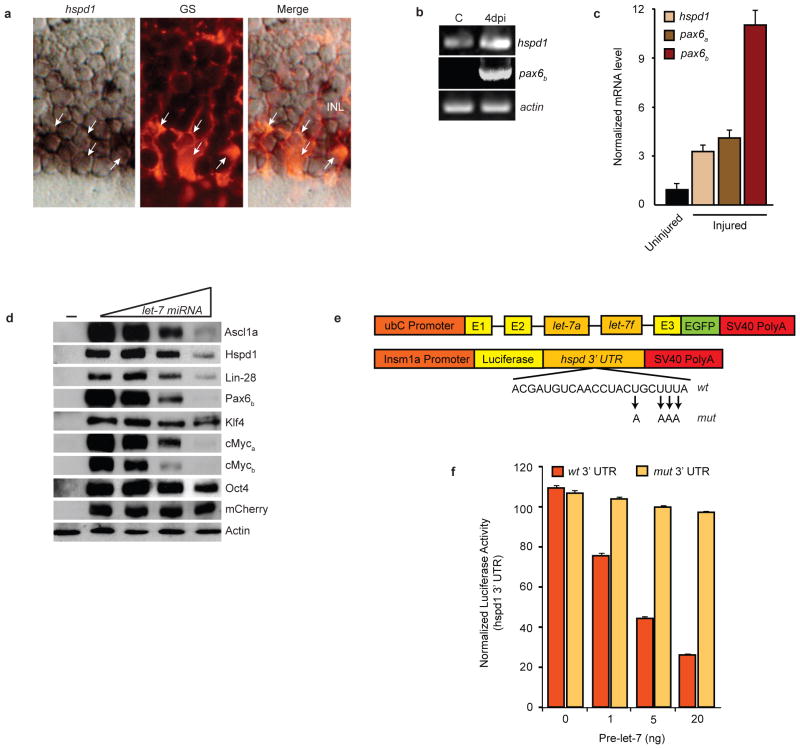
Figure 5.
let-7 miRNAs suppress expression of regeneration and pluripotency-associated genes. (a) hspd1 in situ hybridization and glutamine synthetase (GS) immunofluorescence shows hspd1 RNA co-localizes (arrows) with GS-expressing MG in the uninjured retina. (b) RT-PCR shows hspd1 mRNA expression in uninjured retina that is increased following retinal injury, while pax6b expression is undetectable in the uninjured retina and dramatically induced following retinal injury. (c) Real-time PCR quantification of hspd1, pax6a and pax6b mRNA levels in uninjured and 4 day post injured retina (injured). Data represent means ± s.d. (n=3 fish; compared to uninjured retina, P=0.0001 or less for hspd1, pax6a and pax6b, respectively, at 4 dpi. (d) let-7-dependent suppression of zebrafish proteins that are necessary for retina regeneration or associated with pluripotency. HEK 293 cells were transfected with flag or myc-tagged regeneration or pluripotency-associated gene expression vectors (50 ng) with increasing concentrations (0, 50, 200, 500 ng) ubC:let-7a,let-7f along with pCS2:mCherry for normalization. (−) lane indicates untransfected cells. Forty-eight hrs later cells were harvested and proteins resolved by denaturing SDS-PAGE. Western blots were probed with anti-mCherry, anti-Flag or anti-Myc antibodies. let-7-dependent regulation of protein expression was quantified by densitometry (Supplementary Information, Fig. S7). Experiments were repeated 3 times with similar results. See Fig. S9 for full scans. (e) Constructs used to overexpress let-7 and investigate the function of putative let-7 miRNA binding sites in the hspd1 3’ UTR. Mutations in the let-7 binding site seed sequence are indicated. (f) The hspd1 3’ UTR let-7 binding site at position 1970 confers let-7-dependent regulation on luciferase expression. Luciferase activity from 24 hpf zebrafish embryos that were co-injected at the single cell stage with the luciferase reporter and the let-7 miRNA expression vector described in (e) along with capped Renilla luciferase mRNA for normalization. Data represent means ± s.d. (n=3 fish; compared to wt 3’UTR construct in absence of pri-let-7 miRNA or mut 3’ UTR construct with pri-let-7 miRNA, P=0.0001 or less for luciferase-wt 3’ UTR activity treated with 1, 5 and 20 ng of ubC:let-7a,let- 7f construct.