Abstract
Taxonomically restricted genes or lineage-specific genes contribute to morphological diversification in metazoans and provide unique functions for particular taxa in adapting to specific environments. To understand how such genes arise and participate in morphological evolution, we have investigated a gene called nematogalectin in Hydra, which has a structural role in the formation of nematocysts, stinging organelles that are unique to the phylum Cnidaria. Nematogalectin is a 28-kDa protein with an N-terminal GlyXY domain (glycine followed by two hydrophobic amino acids), which can form a collagen triple helix, followed by a galactose-binding lectin domain. Alternative splicing of the nematogalectin transcript allows the gene to encode two proteins, nematogalectin A and nematogalectin B. We demonstrate that expression of nematogalectin A and B is mutually exclusive in different nematocyst types: Desmonemes express nematogalectin B, whereas stenoteles and isorhizas express nematogalectin B early in differentiation, followed by nematogalectin A. Like Hydra, the marine hydrozoan Clytia also has two nematogalectin transcripts, which are expressed in different nematocyte types. By comparison, anthozoans have only one nematogalectin gene. Gene phylogeny indicates that tandem duplication of nematogalectin B exons gave rise to nematogalectin A before the divergence of Anthozoa and Medusozoa and that nematogalectin A was subsequently lost in Anthozoa. The emergence of nematogalectin A may have played a role in the morphological diversification of nematocysts in the medusozoan lineage.
Keywords: Cnidaria, evolution, exon duplication, nematocyst tubule
Organisms within a phylum commonly share phenotypic traits that are unique to the phylum and strongly associated with specific adaptations. These phenotypic traits are often associated with taxonomically restricted genes. In Cnidaria, the nematocyst, an explosive organelle used to capture prey or repel predators, is such a phylum-specific trait. All nematocysts consist of two primary structures, a capsule and a tubule (1, 2) (Fig. S1A). Modifications of the nematocyst structure such as the capsule size and the armature of the tubule with spines generate different types of nematocysts. Anthozoans have simple nematocyst types (3), whereas medusozoans (Hydrozoa, Cubozoa, and Scyphozoa) have more complex nematocysts exhibiting various tubule and spine morphologies (4). Hydra, a freshwater hydrozoan, has four types of nematocysts: stenotele, desmoneme, holotrichous isorhiza, and atrichous isorhiza.
Nematocysts are formed in large post-Golgi vacuoles during the differentiation of nematocytes. The capsule wall is formed on the inner surface of the vacuole, which grows in size with continuing input of precursor proteins from the Golgi complex (5). After completion of the capsule, tubule formation begins at the apical end, growing outward as a long extension that pushes the vacuole membrane into the cytoplasm (Fig. S1B). After completion of the external tubule, it invaginates through itself to form the internal tubule (1, 6); at that point, spines are added and the capsule wall hardens. Thereafter, the capsule swells as a result of formation of poly-γ-glutamate in the matrix and the accompanying increase in osmotic pressure (7).
A large number of nematocyte-specific genes have been identified (8, 9), many of which have no ortholog in other metazoans and hence appear to be taxonomically restricted. In this report, we investigate the role of one such gene in nematocyst formation. The gene encodes a unique protein with an unusual domain organization. We have named the protein nematogalectin (or nemgal) to emphasize its occurrence in nematocytes and its sugar-binding galectin domain. There are two nematogalectins, A and B, in Hydra, and they are the products of alternative splicing. We demonstrate mutually exclusive expression of nematogalectin A and B and use immunocytochemistry and immunogold EM to show that nematogalectins are major components of the nematocyst tubule.
Results
Genomic Organization and Domain Architecture of Nematogalectins.
Nematogalectin A and B transcripts are formed by an alternative splicing event (Fig. S2A). Exons 1 and 2 are nontranslated 5′-UTR sequences. Exon 3 encodes a signal peptide that is common to both nematogalectin A and B. Exons 4 and 5 encode the mature nematogalectin B protein, whereas exons 6 and 7 encode the mature nematogalectin A protein. Nematogalectins A and B have 294 and 292 amino acids, respectively (Fig. S2B). The 22 amino acids at the amino terminus represent a signal peptide encoded by exon 3, which is common to both nematogalectins and required for translation on the endoplasmic reticulum. The N-terminal 38 amino acids of the mature proteins are almost identical in nematogalectin A and B and in nematogalectin orthologs in other cnidarians (Fig. S2B; see below). This conserved domain is followed by 30 glycine followed by two hydrophobic amino acids (GlyXY) repeats, which can form a collagen triple helix. Similar GlyXY repeats (12–16 repeats) are found in all minicollagens and have been shown to form short collagen triple helices (5, 10). The C terminus consists of a galactose-binding lectin domain followed by a lysine- and arginine-rich sequence. Mapping the protein domains to the exon–intron structure indicates that the signal peptide, GlyXY and galectin domains are each encoded by a single exon.
Nematogalectin Transcripts Are Expressed in Differentiating Nematocytes.
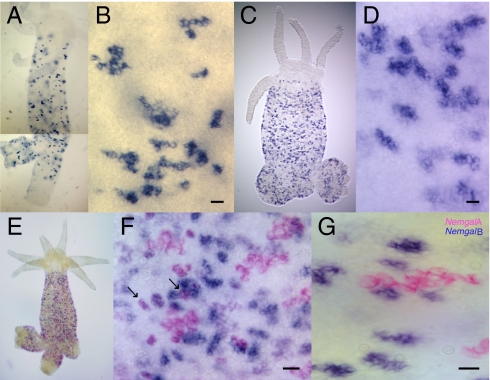
The nematogalectin gene produces four alternatively spliced transcripts: Two encode nematogalectin A, and two encode nematogalectin B. We examined the expression of nematogalectin A and B mRNA by whole-mount in situ hybridization. Nematogalectin A and B probes stained nests of differentiating nematocytes in the ectoderm throughout the gastric region of Hydra (Fig. 1 A–D). The number and morphology of the nests stained by nematogalectin A and B probes appeared to differ: The nematogalectin B probe stained more nests, and most of them were smaller than nests stained by the nematogalectin A probe. This suggested that nematogalectin A and B transcripts are expressed in different nests of nematocytes. Double in situ hybridizations (Fig. 1 E–G) confirmed this idea, because there were almost no nests expressing both nematogalectin A and B transcripts. These results, however, do not rule out the possibility that nematogalectin A and B transcripts are expressed in a temporal sequence during nematocyte differentiation, and thus do not occur simultaneously in the same nests (see below).
Fig. 1.
Whole-mount in situ hybridization of Hydra nematogalectins. Expression of nematogalectin A and B (Nemgal A and Nemgal B) transcripts was detected in differentiating nematocyte nests localized in the gastric region. (A) Two exposures of the same animal were combined for this in situ because the bottom part of one exposure was too dark to show the blue labeled cells clearly. Nematogalectin A antisense probe (A and B), nematogalectin B antisense probe (C and D), and nematogalectin A-expressing (pink) and B-expressing (blue) nematocytes (E–G) are stained with Fast Red and NBT/BCIP, respectively. (F) Arrows indicate coexpression of nematogalectin A and B in some nematocyte nests. (Scale bar: B, D, F, and G, 20 μm.)
To confirm that cells expressing nematogalectin A and B transcripts are differentiating nematocytes, we performed double in situ hybridizations with a minicollagen-1 probe (Fig. S3). All nests of differentiating nematocytes express minicollagen-1 (11), whereas only some nests coexpressed minicollagen-1 and nematogalectin A transcript (Fig. S3C). Similarly, some nests coexpressed minicollagen-1 and nematogalectin B transcript (Fig. S3F). The morphology of stained nests indicated that nests coexpressing a nematogalectin transcript already had a clearly visible vacuole containing the differentiating nematocyst, whereas minicollagen-1 also stained younger nests in which the vacuole was not yet visible. Thus, it appears that nematogalectin A and B transcripts are expressed later in differentiation, after the onset of minicollagen-1 expression.
Structure of Nematogalectin A and B Proteins in Differentiating Nematocytes and Mature Capsules.
The structure of nematogalectin A and B proteins was analyzed with specific antibodies. The nematogalectin B antibody was made against the C-terminal lectin domain and is specific for nematogalectin B (Fig. S4A). The nematogalectin A antibody was made to a truncated nematogalectin A construct containing both GlyXY repeats and the lectin domain (Fig. S4A); it reacts strongly with nematogalectin A but also cross-reacts weakly with nematogalectin B, presumably because of the presence of GlyXY repeats in both proteins (Fig. S4 B and C).
Fig. S5A shows Western blots of lysates prepared from whole Hydra and from mature nematocyst capsules stained with antibodies to nematogalectin A and B. Both Hydra lysates and capsules showed a major band with an apparent molecular weight of 37 kDa with both antibodies. This is higher than the expected molecular weight of 28 kDa based on the amino acid sequence and appears to be attributable to N-glycosylation of nematogalectin. Both nematogalectin A and B, as well as all other known cnidarian homologs (Fig. S2B), have a predicted N-glycosylation site (Asn37) in the highly conserved N-terminal domain. A similar band shift caused by posttranslational glycosylation has been reported for minicollagen-1 (5). A second putative N-glycosylation site is mutated in some of the medusozoan sequences, suggesting that it may not be used in vivo. This idea is further supported by the fact that nematogalectin A and B have the same size in vivo (Fig. S5A).
To localize nematogalectins in nematocyst capsules, we used DTT, a reducing agent, to solubilize the capsule wall (12) and the tubule (13) but not the spines (14). Isolated capsules were treated with DTT and analyzed by Western blotting. Nematogalectin remained in the pellet at low DTT concentrations (0–0.1 mM) but became soluble as the DTT concentration increased (0.5 mM) and was dissolved completely in 2 mM DTT (Fig. S5B). This result indicates that nematogalectin is probably present in the capsule wall or the tubule and not in the spines.
The triple-helix structure of nematogalectin was examined by heat denaturation with or without reducing agent. Mature capsules were isolated from tentacles, incubated with different concentrations of β-mercaptoethanol, and analyzed by SDS/PAGE. In the absence of heat and reducing agent, no nematogalectin was detected in the gel as it formed a complex that was retained in the loading well (lane 1, Fig. S5C). In the presence of a reducing agent, bands at 120 and 37 kDa were observed (lanes 2–5, Fig. S5C), which correspond, respectively, to the triple-helix trimer form and the monomer form of nematogalectin. Treatment of nematocysts at 100 °C for 10 min caused dissociation of capsules and unfolding of the nematogalectin triple helix, giving rise to the nematogalectin monomer with a molecular weight of 37 kDa (lanes 6–10, Fig. S5C).
Tubule Formation During Nematocyst Development.
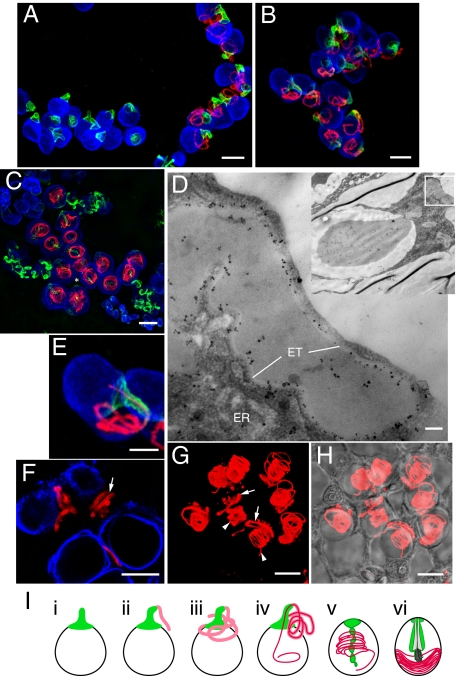
The mutually exclusive expression of nematogalectin A and B transcripts suggested that the two products might fulfill different functions in nematocysts despite their high sequence similarity. To test this idea, we analyzed the localization of nematogalectin A and B in developing nematocysts by confocal microscopy using antibodies to nematogalectin A and nematogalectin B as well as specific antibodies to minicollagen-1 and minicollagen-15. Although the nematogalectin A antibody showed some cross-reactivity with nematogalectin B in Western blots (Fig. S4C), the two antibodies recognized distinct structures and showed essentially no cross-reactivity in immunostaining experiments. Nematogalectin A and B antibodies stained the tubules of stenoteles throughout nematocyst formation (Fig. 2). Based on the patterns of immunostaining, we were able to divide the process of tubule formation into four stages: protrusion, elongation, inversion/coiling, and compression. Nematogalectin B staining first appeared at the onset of tubule formation as a protrusion at the apical end of the developing capsule (Fig. 2A, nest at left). The green fluorescent signal corresponds to the future shaft of a stenotele tubule. Nematogalectin A antibody then stained the rest of tubule when it continued to elongate and wound randomly through the cytoplasm (Fig. 2A, nest at right). This staining pattern indicates that nematogalectin A and B are sequentially expressed during stenotele differentiation in agreement with the in situ hybridization results (Fig. 1).
Fig. 2.
Immunostaining of developing stenotele nematocysts with nematogalectin A and B antibodies. (A–C) 3D confocal images of differentiating stenoteles showing distinct localization of nematogalectin B (green) at the base of the external tubule, which later forms the shaft, and nematogalectin A (red) along the distal tubule. The stenotele nest in C is marked by an asterisk. (D) Transmission EM of immunogold-labeled nematogalectin A showing the localization of gold particles over the wall of the external tubule and over the endoplasmic reticulum. ER, endoplasmic reticulum; ET, wall of external tubule. (E) Enlarged view of a stenotele (from B) showing a distal tubule (red) invaginating through the proximal tubule (green). (F) Single confocal section through the external tubule showing the presence of nematogalectin A in tubule wall (arrow). (G) Fluorescent image of nematogalectin A-stained stenoteles. The arrows and arrowheads indicate the external tubule and inverted tubule, respectively. (H) Merged fluorescent image in G and bright-field image. (I) Schematic drawings depicting the process of tubule development: protrusion (i), elongation (ii and iii), inversion/coiling (iv and v), and compression (vi). Red signal, nematogalectin A; green signal, nematogalectin B; blue signal, minicollagen-1. (Scale bars: A–C and F–H, 10 μm; D, 0.1 μm; E, 5 μm.)
To localize nematogalectin A within the developing tubule, we performed immunogold EM. The results showed that the gold particles were predominantly localized over the wall of the external tubule as well as in the endoplasmic reticulum (Fig. 2D). Single confocal sections also demonstrated that the nematogalectin A antibody stained the wall of the external tubule (Fig. 2F). After tubule growth stopped, the tubule inverted through itself into the capsule. This process is called inversion and occurs rapidly (2, 6, 15) (Fig. 2 B and E). The tubule diameter before inversion (Fig. 2A) was reduced to two-thirds of its original size after inversion (Fig. 2C). Although inversion is a rapid process, we observed some differentiating stenoteles in which part of the tubule had entered the capsule and had a small diameter, whereas a small portion of the tubule still remained outside the capsule with a larger diameter (Fig. 2 E, G, and H). Once the tubule had completely entered the capsule, it was coiled throughout the capsule lumen (Fig. 2C). At this stage, the spines of the stenotele begin to form inside the proximal part of the inverted tubule (14). As they grow in size, this part of the tubule expands and pushes the coils of the thin distal tubule to the edge of the capsule, a process that we call compression. Nematogalectin A staining becomes weaker at this stage (Fig. 3D), probably as a result of compaction of the tubule wall. In discharged stenoteles, nematogalectin A staining was again visible throughout the tubule, with the fluorescence signal increasing from proximal to distal along the tubule (Fig. S6). Fig. 2I summarizes the steps in stenotele tubule formation schematically.
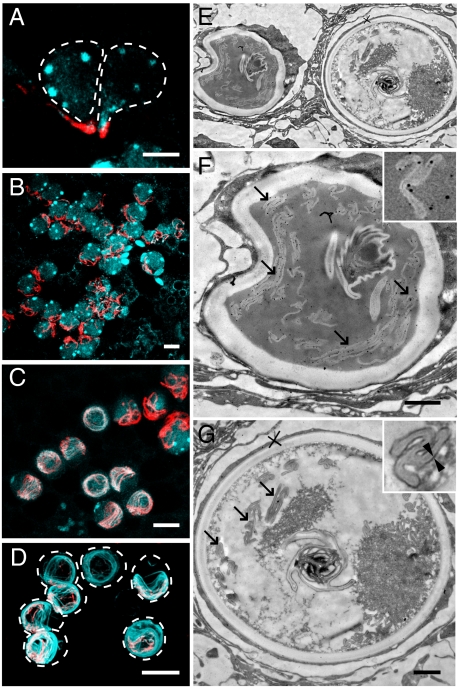
Fig. 3.
Immunostaining of developing stenotele nematocysts with nematogalectin A and minicollagen-15 antibodies. (A–D) Nematogalectin A (red) and minicollagen-15 (light blue) antibodies stain nematocyte nests at various stages of differentiation. Colocalization of nematogalectin A and minicollagen-15 on the tubule is only observed at later stages, as shown in C and D. (E–G) Transmission EM images of immunogold-labeled sections. (F) Enlarged image of late-stage differentiating stenotele in E. Gold particles are visible over the invaginated tubule (arrows). (Inset) Higher magnification. (G) Enlarged image of mature stenotele in E. No gold labeling is present over the invaginated tubule (arrows). (Inset) Outer layer of the tubule is coated with additional dark material at this stage (two arrowheads). (Scale bars: A, 5 μm; B–D, 10 μm; F and G, 1 μm.)
Minicollagen-15 is an unusual minicollagen specifically localized to tubules and not present in the wall of the nematocyst capsule (13). Unexpectedly, minicollagen-15 is only colocalized with nematogalectin A at the final stage of tubule development (Fig. 3). During tubule formation, when nematogalectin A is localized in the elongating external tubule (Fig. 2A), minicollagen-15 was detected as dense globules in the capsule matrix (Fig. 3A). It remained in the form of dense globules throughout the process of tubule elongation (Fig. 3B). Not until after the external tubule inverted into the capsule did minicollagen-15 become incorporated into the tubule and colocalize with nematogalectin A (Fig. 3C). The two stenotele nests in Fig. 3C show particularly clear examples of the difference in minicollagen-15 localization before (Upper Right) and after (Center) tubule inversion. Fig. 3E shows a transmission EM image of immature and mature stenoteles lying next to each other. The capsule of the immature stenotele (Fig. 3F) has a distorted shape (incomplete hardening of capsule wall causes capsule distortion during fixation) and is equivalent to the developmental stage shown in Fig. 3C. Immunogold-labeled nematogalectin A (indicated by black dots) could be detected at and near the inverted tubule (arrows in Fig. 3F) at this stage. By comparison, the tubule of the mature stenotele (equivalent to Fig. 3D) was not labeled by immunogold particles (arrows in Fig. 3G). The mature tubule contains an additional layer, which can be seen clearly between two arrowheads (Fig. 3G Inset), and this outer layer appears as a 12-nm-wide region of darker contrast in the image. It seems that immunogold-labeled antibodies no longer react with the tubule after the formation of this dark layer.
The structures of atrichous and holotrichous isorhizas are quite similar during early stages of tubule outgrowth. Hence, to analyze developing atrichous isorhizas clearly, we used a mutant strain, nem-3, which lacks holotrichous isorhizas (16). Fig. S7 A and B shows that nematogalectin B was expressed at the onset of tubule development in atrichous isorhizas, forming the proximal part of the tubule. Nematogalectin A was expressed later and formed the distal part of the elongating tubule (Fig. S7 C and D). In WT Hydra (strain 105), there were developing holotrichous isorhizas in addition to developing atrichous isorhizas. These were somewhat larger than atrichous isorhizas (Fig. S7 E and F) but also expressed nematogalectin B in the proximal tubule and nematogalectin A in the distal tubule. Fig. S7F shows an inversion stage with the red distal tubule invaginating through the green proximal tubule. The inverted tubule in holotrichous isorhizas forms a clear spiral at one end of the capsule. Desmonemes, in contrast to stenoteles and isorhizas, express only nematogalectin B. Antibodies against nematogalectin B but not nematogalectin A stained the developing tubules of desmonemes from the protrusion stage until the completion of the inversion process (Fig. S7 H and I). The external tubule is curved and appears slightly distended (Fig. S7H). After inversion, the tubule is thinner and forms a tight loop within the capsule wall (Fig. S7I).
Nematogalectin in the Marine Hydrozoan Clytia.
To investigate whether the cell type-specific expression of nematogalectin A and B transcripts also occurs in other hydrozoans, we examined the expression of Clytia orthologs (Ch-nemgal A and Ch-nemgal B) in differentiating tentacle bulbs. Expression of both Ch-nemgal A and B transcripts was detected in the tentacle bulbs (Fig. S8 A–D), where nematocytes proliferate and differentiate (17). Weaker signals were also found at the margin of the mouth, where the population of differentiating nematocytes is less dense than in the tentacle bulb. Double in situ hybridization showed two distinct populations of differentiating nematocytes (Fig. S8 E and F) in the tentacle bulbs, with one expressing only Ch-nemgal B transcript (red) and the other expressing both Ch-nemgal A and B transcripts (purple). Expression of Ch-nemgal A and B transcripts was not completely mutually exclusive. Rather, Ch-nemgal A transcript appeared to be expressed in a subpopulation of Ch-nemgal B–expressing nematocytes. Currently, we are not sure if this small population represents a particular type of nematocyte or a specific stage of differentiation. However, it seems clear that Ch-nemgal A and B represent alternatively spliced transcripts, because, like the Hydra genes, they have identical signal peptides and similar but not identical mature protein sequences (Fig. S2B).
Nematogalectins Are Highly Conserved in Cnidarians.
To understand the evolution of nematogalectins, we searched for homologous sequences in other cnidarians. Nematogalectin A and B sequences were identified in EST libraries from Hydra viridis, Hydra oligactis, and Hydra vulgaris AEP (18) and in the marine hydrozoan Clytia (see above). By contrast, only nematogalectin B orthologs could be found in EST and genomic data [GenBank, DNA Data Base in Japan (DDBJ), and European Molecular Biology Laboratory (EMBL)] available for the anthozoans Nematostella, Acropora, Metridium, Anemonia, and Aiptasia. The domain organization in all nematogalectin sequences is well conserved. Furthermore, sequence alignment shows that the N-terminal 38 amino acids are almost identical in all nematogalectin sequences (Fig. S2B), although the anthozoan and medusozoan lineages have been evolving independently for more than 550 million years (19, 20). The function of this remarkably conserved domain is presently unknown.
In addition to nematogalectin A and B, we identified two more sequences in the Hydra genome that contain a signal peptide, GlyXY repeats, and a galectin domain. We have called these sequences “nematogalectin-related” (Fig. S9). The two nematogalectin-related proteins use different promoters and have different signal peptide exons (only one is shown in Fig. S9), but the same two exons encode the mature nematogalectin-related protein. We also found nematogalectin-related orthologs in ESTs from several anthozoans, the scyphozoan Aurelia, and the hydrozoans Clytia and Hydra (Fig. S9). As in nematogalectins, there is a region of near-sequence identity at the N terminus of all nematogalectin-related orthologs. The C-terminal 30 amino acids are also almost identical in anthozoan and hydrozoan nematogalectin-related sequences but are different from the corresponding sequence in nematogalectins. In situ hybridization has shown that the Hydra (Fig. S10) and coral orthologs (21) of nematogalectin-related are expressed in differentiating nematocytes, indicating that nematogalectin-related is also a nematocyte-specific gene.
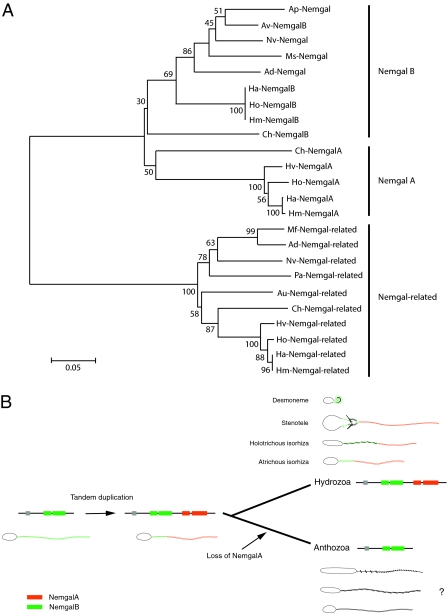
Nematogalectin and nematogalectin-related proteins have the same exon-intron structure and appear to have evolved from an ancient gene duplication at the base of the cnidarian radiation. In nearest neighbor phylogeny (Fig. 4A), nematogalectin and nematogalectin-related proteins form distinct families, each of which contains anthozoan and hydrozoan representatives. The tree topology of the nematogalectin family indicates that there has been a further gene duplication giving rise to nematogalectin A and B sequences. Nematogalectin B sequences are present in both anthozoans and the hydrozoans Hydra and Clytia, whereas nematogalectin A sequences are found only in the hydrozoans Hydra and Clytia. The simplest explanation for this phylogeny is that the nematogalectin gene duplication occurred before the divergence of anthozoans and hydrozoans and the nematogalectin A gene was lost in the anthozoan lineage (“birth and death” model) (22). The occurrence of nematogalectin A and B homologs in the hydrozoan Clytia is consistent with this interpretation. However, the Clytia nematogalectin sequences are remarkably different from the corresponding nematogalectin A and B clusters, and they appear to have diverged early in the phylogenetic tree. Clytia nematogalectins could represent independent duplication and loss events, but wider sequence sampling is needed to confirm this theory.
Fig. 4.
(A) Neighbor-joining tree inferred from nematogalectin and nematogalectin-related sequences. Ad, Acropora millepora; Ap, Aiptasia pallida; Au, Aurelia aurita; Av, Anemonia viridis; Ch, Clytia hemisphaerica; Ha, Hydra vulgaris; Hm, Hydra magnipapillata; Ho, Hydra oligactis; Hv, Hydra viridis; Mf, Montastraea faveolata; Ms, Metridium senile; Nv, Nematostella vectensis; Pa, Porites astreoides. (B) Schematic drawing of the nematogalectin locus showing tandem duplication of nemgal B exons (green) to yield nemgal A exons (red). Exon encoding the signal peptide is shown in the gray box. The nemgal A duplicate was subsequently lost in the anthozoan lineage. The localization of nemgal A (red) and nemgal B (green) proteins is shown in tubules of desmoneme, stenotele, holotrichous, and atrichous isorhizas. The localization of nematogalectin in anthozoan nematocysts is presently not known.
Discussion
Nematogalectin Is a Major Component of the Nematocyst Tubule.
Our results extend earlier EM observations on tubule formation (6, 15, 23), with additional information on the molecules involved. We have identified nematogalectin as a major component of the nematocyst tubule and shown that it is incorporated into the external tubule during outgrowth from the capsule and is also present in the final structure after tubule inversion. This is in contrast to a second tubule-specific protein, minicollagen-15, which accumulates in globules in the matrix of the developing capsule before attaching to the surface of the tubule after it has invaginated (13) (Fig. 3 C and D). This suggests that nematogalectin could act as a substrate for addition of minicollagen-15.
It is not clear what holds the tubule together as it forms. The presence of chondroitin in proteoglycan on the inner surface of the vacuolar membrane surrounding the elongating tubule (24) suggests that it could be the substrate onto which nematogalectin binds during outgrowth of the external tubule. This agrees with the localization of nematogalectin in transmission EM images, where it overlaps the homogeneous tubule wall structure (Fig. 2D). Because nematogalectin is a 120-kDa trimer and has three galactose-binding domains, it could bind to multiple chondroitin glycosaminoglycan molecules and stabilize the chondroitin proteoglycan layer. This appears to be necessary because this structure behaves as a molecular sheet or “tissue” that inverts through itself during formation of the internal tubule. Following inversion, minicollagen-15 assembles on the nematogalectin layer (Fig. 3 C and D) to form the final stable tubule structure. The highly conserved N- and C-terminal sequences in nematogalectins may be involved in the interaction with minicollagen.
Alternative Splicing of Nematogalectin Generates Cell-Specific Transcripts.
Alternative splicing generates more than one transcript from a single gene, and these alternative transcripts can be expressed in different cell types or tissues, leading to enhanced protein diversity and functional complexity (25). The data presented here demonstrate that alternative splicing generates two nematogalectin transcripts and that these are mutually exclusively expressed, either in different nematocyte types or in one type at different times during differentiation. A similar example of alternative splicing was recently described in Hydra minicollagens (4). Minicollagen genes are grouped into clusters, each containing three genes formed by tandem duplication of the exon encoding the mature minicollagen peptide. Alternative splicing links the common signal peptide exon in each cluster to one of the three minicollagen exons.
Nematogalectin A and B sequences in Hydra are the result of tandem duplication of the two exons encoding the mature protein sequence (Fig. S2A). Both proteins use the same upstream exon encoding the signal peptide sequence. Clytia, like Hydra, has two different nematogalectin transcripts, and they encode identical signal peptide sequences but different mature protein sequences. Although the genome sequence of Clytia is not known, this situation suggests that the tandem exon duplication is also present in Clytia and that the two Clytia nemtogalectin transcripts are the result of alternative splicing.
Tandem exon duplication events can yield equivalent protein sequences if the duplication event is accompanied by mutually exclusive alternative splicing. Letunic et al. (26) and Kondrashov and Koonin (27) have found that the vast majority of genes arising from exon duplication in human, fly, and worm genomes are involved in mutually exclusive alternative splicing. Our finding of mutually exclusive alternative splicing in Hydra argues that this process dates back at least to the common ancestor of cnidarians and bilaterians.
Nematocyst Evolution and the Origin of Different Nematogalectin Sequences.
The external tubule, which is everted when capsules explode, is the “business end” of nematocysts and is correspondingly different in different nematocyst types. All cnidarians have the so-called “atrichous” type of nematocyst, and these are thought to represent the primordial nematocyst type (4, 28, 29). As cnidarians evolved within the anthozoan and medusozoan lineages, a variety of additional nematocyst types with increasingly complex tubule structures appeared, such as euryteles, stenoteles, and desmonemes (4). The molecular basis for the diversity of tubule structures is presently unknown; thus far, only three tubule-specific proteins have been identified: nematogalectin A and B described here, minicollagen-15 (13), and spinalin (14). Although it seems clear that more proteins are involved in tubule morphogenesis, our results permit some speculation about the mode of tubule evolution.
Nematogalectin A arose by tandem duplication of nematogalectin B. The phylogeny in Fig. 4A is consistent with exon duplication before the separation of the anthozoan and medusazoan lineages, followed by loss of nematogalectin A in the anthozoan lineage (22) (Fig. 4B). As a consequence of mutually exclusive alternative splicing, nematogalectin A is not coexpressed with nematogalectin B. During nematocyst formation, nematogalectin B is expressed first and is localized in the proximal tubule, whereas nematogalectin A is expressed later and is localized in the distal tubule. Hence, the two nematogalectin proteins define different parts of the tubule and could provide a substrate for the evolution of unique functions and/or structures in the tubule. For example, the proximal tubule expresses nematogalectin B and has a high density of large spines or spine ridges (Fig. S1 C and D). By comparison, the distal tubule of the three nematocyst types expressing nematogalectin A is thinner and lacks spines completely or has a low density of spines (Fig. S1 C and D). The tubule of desmonemes is consistent with this interpretation because it is covered with spines along its whole length and only expresses nematogalectin B and not nematogalectin A. Finally, it is noteworthy that EM images of atrichous nematocysts in anthozoans have shown that they also have spines (30), consistent with the fact that anthozoans have the nemtogalectin B gene but lack the nematogalectin A gene.
The situation with minicollagen-15 (13) is similar to that with nematogalectin. It also evolved as a result of tandem exon duplication and is in a cluster of minicollagen exons, which are alternatively spliced to a common signal peptide exon (4). Taken together, the results on nematogalectins and minicollagen-15 support the idea that the more complex nematocyst repertoire in medusozoans compared with anthozoans is attributable, in part, to a diversification of tubule proteins by tandem exon duplication. The bacterial flagellum and the eukaryotic cilium are similarly complex structures, which, like the nematocyst, appear to have evolved by a series of gene duplication events and associated sequence evolution to yield a variety of proteins from a limited number of starting domain motifs (31, 32).
Materials and Methods
Cloning and Sequencing.
To clone the 5′-nontranslated sequence of nematogalectin A and B transcripts, Hydra cDNAs prepared using the SMART RACE cDNA Amplification Kit (Clontech) were amplified with Advantage 2 PCR polymerase (Clontech) using universal primer mix (UPM) and either gene-specific primer for the nematogalectin A transcript (5′-GGGGCTCCCATAAAACCTTGTGGTCCTGGTGGTCCCTG-3′) or gene-specific primer for the nematogalectin B transcript (5′-CCCTCGCAAATGACAGTGAAGTTGGGCATGATAGCTTCGGG-3′). The subsequent PCR products were cloned directly into the vector pCR4-TOPO (Invitrogen). For each nematogalectin transcript, four plasmids containing the PCR inserts were sequenced.
Expression Pattern of Nematogalectin mRNAs.
Procedures used for whole-mount in situ hybridization are described in SI Text.
Antibodies and Western Blot Analyses.
Construction of recombinant nematogalectin proteins, their specific antibodies, and Western blot analyses are described in SI Text.
Immunofluorescence and Immunogold EM.
Both immunofluorescence EM and postembedding immunogold EM were performed essentially as described by Hwang et al. (33). More details are provided in SI Text.
Sequence Alignment and Phylogenetic Analysis.
Amino acid sequences of nematogalectin or nematogalectin-related homologs were compiled from EST and genomic databases at the GenBank, DDBJ, or EMBL. EST sequences from H. viridis, H. oligactis, and H. vulgaris AEP and Aurelia aurita were provided by Thomas Bosch, Kostya Kalturin, and Georg Hemmrich (Kiel University, Kiel, Germany). A molecular phylogenetic tree of nematogalectin and nematogalectin-related homologs was constructed with the neighbor-joining method with p-distance using MEGA software (34). In brief, 23 sequences, including Hydra nematogalectin A and B, were aligned using the program CLUSTALW (Gonnet protein weight matrix, default pairwise alignment, and 3.0 gap opening/1.8 extension penalties of multiple alignment). Using MEGA, the branch reliability was evaluated with 2,000 bootstrap replications.
Supplementary Material
Acknowledgments
We thank Chie Iwamoto for the construction of recombinant proteins and Akemi Mizuguchi for her assistance in whole-mount in situ hybridization and sequencing.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1003256107/-/DCSupplemental.
References
- 1.Slautterback DB. Nematocyst development. In: Lenhoff HM, Loomis WF, editors. The Biology of Hydra and of Some Other Coelenterates. FL: Univ of Miami Press; 1961. pp. 77–130. [Google Scholar]
- 2.Watson GM. Ultrastructure and cytochemistry of developing nematocysts. In: Hessinger DA, Lenhoff HM, editors. The Biology of Nematocysts. New York: Academic; 1988. pp. 143–164. [Google Scholar]
- 3.Cutress CE. An interpretation of the structure and distribution of cnidae in Anthozoa. Syst Biol. 1955;4:120–137. [Google Scholar]
- 4.David CN, et al. Evolution of complex structures: Minicollagens shape the cnidarian nematocyst. Trends Genet. 2008;24:431–438. doi: 10.1016/j.tig.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Engel U, et al. A switch in disulfide linkage during minicollagen assembly in Hydra nematocysts. EMBO J. 2001;20:3063–3073. doi: 10.1093/emboj/20.12.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holstein T. The morphogenesis of nematocysts in Hydra and Forskålia: An ultrastructural study. J Ultrastruct Res. 1981;75:276–290. doi: 10.1016/s0022-5320(81)80085-8. [DOI] [PubMed] [Google Scholar]
- 7.Szczepanek S, Cikala M, David CN. Poly-γ-glutamate synthesis during formation of nematocyst capsules in Hydra. J Cell Sci. 2002;115:745–751. doi: 10.1242/jcs.115.4.745. [DOI] [PubMed] [Google Scholar]
- 8.Hwang JS, et al. The evolutionary emergence of cell type-specific genes inferred from the gene expression analysis of Hydra. Proc Natl Acad Sci USA. 2007;104:14735–14740. doi: 10.1073/pnas.0703331104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Milde S, et al. Characterization of taxonomically restricted genes in a phylum-restricted cell type. Genome Biol. 2009;10:R8. doi: 10.1186/gb-2009-10-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Özbek S, et al. Structure/function relationships in the minicollagen of Hydra nematocysts. J Biol Chem. 2002;277:49200–49204. doi: 10.1074/jbc.M209401200. [DOI] [PubMed] [Google Scholar]
- 11.Kurz EM, Holstein TW, Petri BM, Engel J, David CN. Mini-collagens in hydra nematocytes. J Cell Biol. 1991;115:1159–1169. doi: 10.1083/jcb.115.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Özbek S, et al. The glycoprotein NOWA and minicollagens are part of a disulfide-linked polymer that forms the cnidarian nematocyst wall. J Biol Chem. 2004;279:52016–52023. doi: 10.1074/jbc.M407613200. [DOI] [PubMed] [Google Scholar]
- 13.Adamczyk P, et al. Minicollagen-15, a novel minicollagen isolated from Hydra, forms tubule structures in nematocysts. J Mol Biol. 2008;376:1008–1020. doi: 10.1016/j.jmb.2007.10.090. [DOI] [PubMed] [Google Scholar]
- 14.Koch AW, et al. Spinalin, a new glycine- and histidine-rich protein in spines of Hydra nematocysts. J Cell Sci. 1998;111:1545–1554. doi: 10.1242/jcs.111.11.1545. [DOI] [PubMed] [Google Scholar]
- 15.Skaer RJ. The secretion and development of nematocysts in a siphonophore. J Cell Sci. 1973;13:371–393. doi: 10.1242/jcs.13.2.371. [DOI] [PubMed] [Google Scholar]
- 16.Sugiyama T, Fujisawa T. Genetic analysis of developmental mechanisms in hydra. I. Sexual reproduction of Hydra magnipapillata and isolation of mutant. Dev Growth Differ. 1977;19:187–200. doi: 10.1111/j.1440-169X.1977.00187.x. [DOI] [PubMed] [Google Scholar]
- 17.Denker E, Manuel M, Leclère L, Le Guyader H, Rabet N. Ordered progression of nematogenesis from stem cells through differentiation stages in the tentacle bulb of Clytia hemisphaerica (Hydrozoa, Cnidaria) Dev Biol. 2008;315:99–113. doi: 10.1016/j.ydbio.2007.12.023. [DOI] [PubMed] [Google Scholar]
- 18.Khalturin K, et al. A novel gene family controls species-specific morphological traits in Hydra. PLoS Biol. 2008;6:e278. doi: 10.1371/journal.pbio.0060278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Putnam NH, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 20.Chapman JA, et al. The dynamic genome of Hydra. Nature. 2010;464:592–596. doi: 10.1038/nature08830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grasso LC, et al. Microarray analysis identifies candidate genes for key roles in coral development. BMC Genomics. 2008;9:540–547. doi: 10.1186/1471-2164-9-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slautterback DB, Fawcett DW. The development of the cnidoblasts of Hydra; an electron microscope study of cell differentiation. J Biophys Biochem Cytol. 1959;5:441–452. doi: 10.1083/jcb.5.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adamczyk P, et al. A non-sulfated chondroitin stabilizes membrane tubulation in cnidarian organelles. J Biol Chem. 2010;285:25613–25623. doi: 10.1074/jbc.M110.107904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graveley BR. Alternative splicing: Increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 26.Letunic I, Copley RR, Bork P. Common exon duplication in animals and its role in alternative splicing. Hum Mol Genet. 2002;11:1561–1567. doi: 10.1093/hmg/11.13.1561. [DOI] [PubMed] [Google Scholar]
- 27.Kondrashov FA, Koonin EV. Origin of alternative splicing by tandem exon duplication. Hum Mol Genet. 2001;10:2661–2669. doi: 10.1093/hmg/10.23.2661. [DOI] [PubMed] [Google Scholar]
- 28.Weill R. Contribution à l’étude des cnidaires et le leurs nematocystes. I. Trav Sta Zool Wimereux. 1934;10:1–347. [Google Scholar]
- 29.Werner B. New investigations on systematics and evolution of the class Scyphozoa and the phylum Cnidaria. Publ Seto Mar Biol Lab. 1973;20:35–61. [Google Scholar]
- 30.Schmidt H. Die Nesselkapseln der Anthozoen und ihre Bedeutung für die phylogenetische Systematik. Helgoland Marine Res. 1972;23:422–458. [Google Scholar]
- 31.Jékely G, Arendt D. Evolution of intraflagellar transport from coated vesicles and autogenous origin of the eukaryotic cilium. Bioessays. 2006;28:191–198. doi: 10.1002/bies.20369. [DOI] [PubMed] [Google Scholar]
- 32.Liu R, Ochman H. Stepwise formation of the bacterial flagellar system. Proc Natl Acad Sci USA. 2007;104:7116–7121. doi: 10.1073/pnas.0700266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hwang JS, et al. Cilium evolution: Identification of a novel protein, nematocilin, in the mechanosensory cilium of Hydra nematocytes. Mol Biol Evol. 2008;25:2009–2017. doi: 10.1093/molbev/msn154. [DOI] [PubMed] [Google Scholar]
- 34.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.