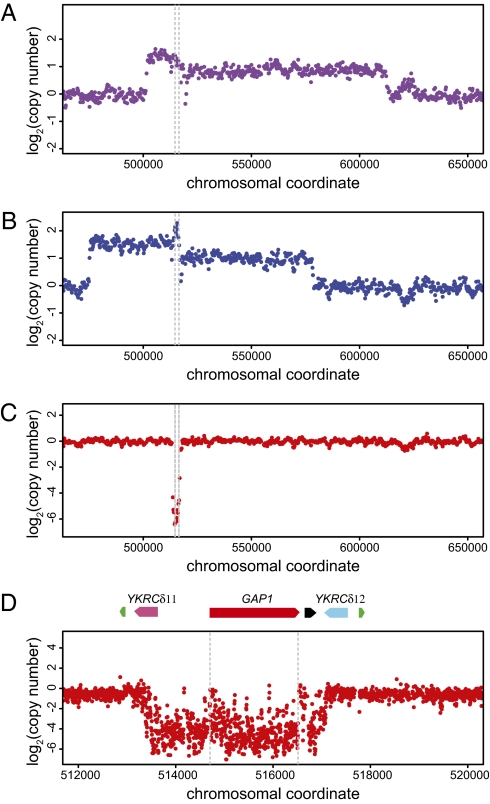
Fig. 1.
Gene amplification and deletion alleles at the GAP1 locus identified by array comparative genomic hybridization. We identified frequent copy-number polymorphisms at the GAP1 locus in clones subjected to long-term adaptation in diverse nitrogen-limited chemostats. These included unique amplification alleles in clones adapted to l-glutamine (A) and l-glutamate (B). Each of these amplification alleles is ∼100 kb and includes GAP1 (bounded by gray dashed lines in all plots) and several flanking genes. In addition, we identified deletion events that include only the GAP1 gene in several clones, including one adapted to allantoin limitation (C). We hybridized DNA from the allantoin-adapted clone to an overlapping tiling microarray (D) and determined that the deletion event is bounded by two flanking long terminal repeats: YKRCδ11 and YKRCδ12. The locus also includes an origin of replication (black) and two tRNA genes (green) that lie distal to the LTRs.