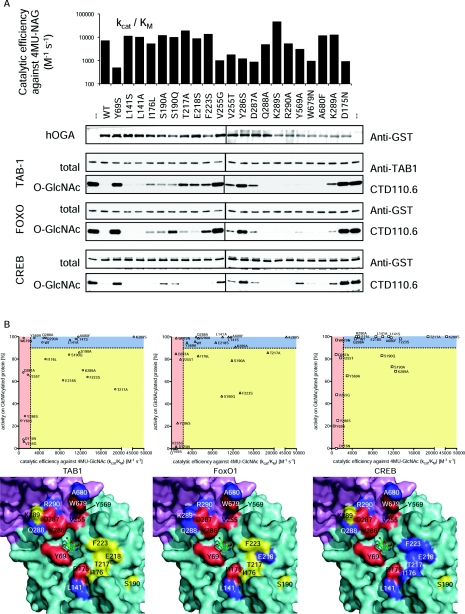
Figure 3. Activity of hOGA groove mutants.
Twenty individual mutants of hOGA were generated based on the structural information obtained from OgOGA. (A) To evaluate the structural integrity of the hOGA mutants, the catalytic efficiency of 4MU-GlcNAc turnover was determined and compared with the wild-type (kcat/Km=7000 M−1·s−1) and the catalytically impaired D175N mutant (negative control). hOGA mutants were also incubated with O-GlcNAc substrate proteins TAB1, FoxO1 and CREB, and deglycosylation was observed by Western blotting. (B) Visualization of the correlation between the mutants' ability to hydrolyse the pseudosubstrate 4MU-GlcNAc and glycoprotein substrates. Catalytically impaired mutants (kcat/Km for 4MU-GlcNAc ≤50% of wild-type) locate in the areas shaded in red. Mutants unaffected in both 4MU-GlcNAc turnover and protein deglycosylation (cut-off 10% reduction of wild-type activity) are shown in the blue areas. Mutations reducing OGA activity on protein substrates only are depicted in the yellow areas. Location of the mutations is shown on the surface around the active site of OgOGA (coloured by domain, see Figure 2A). The mutated amino acids are shaded according to correlation plots, thus amino acids affecting protein substrate recognition are highlighted in yellow.