Abstract
Diffuse large B-cell lymphoma (DLBCL), the most common form of lymphoma in adulthood, comprises multiple biologically and clinically distinct subtypes including germinal center B cell-like (GCB) and activated B cell-like (ABC) DLBCL1. Gene expression profile studies have shown that its most aggressive subtype, ABC-DLBCL, is associated with constitutive activation of the NF-kB transcription complex2. However, except for a small fraction of cases3, it remains unclear whether NF-kB activation in these tumors represents an intrinsic program of the tumor cell of origin or a pathogenetic event. Here we show that >50% of ABC-DLBCL and a smaller fraction of GCB-DLBCL carry somatic mutations in multiple genes, including negative (TNFAIP3/A20) and positive (CARD11, TRAF2, TRAF5, MAP3K7/TAK1 and TNFRSF11A/RANK) regulators of NF-kB. Of these, the A20 gene, which encodes for a ubiquitin-modifying enzyme involved in termination of NF-kB responses, is most commonly affected, with ~30% of patients displaying biallelic inactivation by mutations and/or deletions. When reintroduced in cell lines carrying biallelic inactivation of the gene, A20 induced apoptosis and cell growth arrest, indicating a tumor suppressor role. Less frequently, missense mutations of TRAF2 and CARD11 produce molecules with significantly enhanced ability to activate NF-kB. Thus, our results demonstrate that NF-kB activation in DLBCL is caused by genetic lesions affecting multiple genes, whose loss or activation may promote lymphomagenesis by leading to abnormally prolonged NF-kB responses.
DLBCL represents a heterogeneous disease in terms of genetic, phenotypic and clinical features. Accordingly, genome-wide expression profile (GEP) studies revealed the existence of several DLBCL categories, reflecting their origin from discrete B cell differentiation stages1 or the co-regulated expression of comprehensive transcriptional signatures4. The cell-of-origin classification schema comprises GCB-DLBCL, derived from a GC centroblast; the less curable ABC-DLBCL, whose expression pattern resembles that of cells committed to plasmacytic differentiation; primary mediastinal large B cell lymphomas (PMBL), arising from thymic B-cells5; and cases that remain unclassified1,6. A key feature of ABC-DLBCL is the activation of the NF-kB signaling pathway, as evidenced by the preferential expression of known NF-kB target genes and the dependence of ABC-DLBCL cell lines on NF-kB activity for proliferation and survival2,7. A recent study reported that ~8% of ABC-DLBCL carry oncogenic mutations of CARD113, a cytoplasmic scaffolding protein required for activation of NF-kB during antigen-dependent signaling8. However, the molecular mechanism underlying NF-kB activation in the remaining large fraction of cases remains unknown, leaving open the possibility that it may reflect a physiologic status of the normal ABC-DLBCL counterpart.
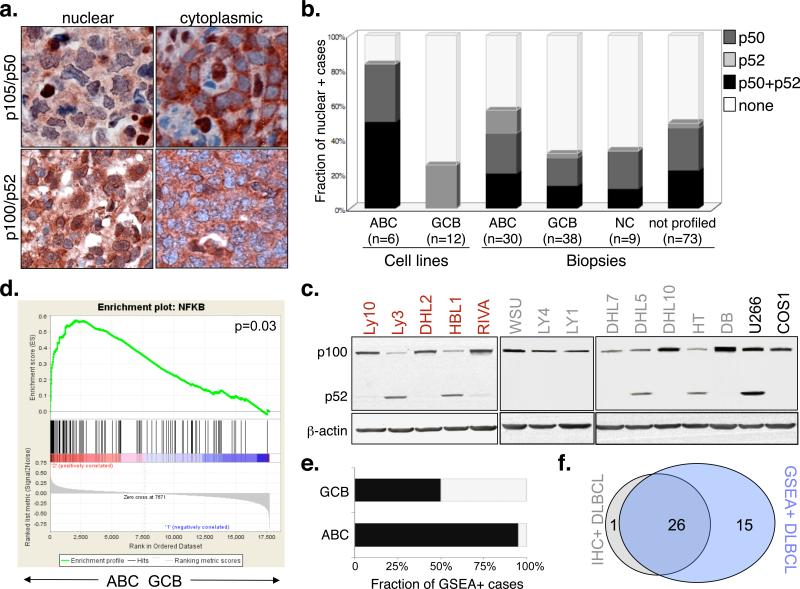
To address this issue, we first characterized 168 DLBCL samples, representative of major subtypes, for the presence of active NF-kB complexes by using immunohistochemical assays detecting nuclear NFKB1/p50 (read-out for the classical pathway) and NFKB2/p52 (alternative pathway)9,10(Figure 1a). Nuclear localization of NF-kB was observed in the tumor cells of 61% ABC-DLBCL and a smaller fraction (30%) of GCB-DLBCL, as well as in 3/9 unclassified and 36/73 not profiled DLBCL (Figure 1b). Both classical and alternative NF-kB pathways were found to be involved, occasionally within the same sample (one third of the positive cases), and consistent with the established role of specific signals (e.g., CD40-CD40L) in the activation of both pathways11,12. Engagement of the alternative NF-kB pathway was also documented by detection of p52, the active product of p100 processing, in Western blot assays (Figure 1c). Gene set enrichment analysis (GSEA) of transcriptionally profiled cases confirmed that the gene expression signature of ABC-DLBCL is significantly enriched in NF-kB target genes (Table S1) with respect to both normal GC centroblasts, used as negative control13 (p<0.005)(not shown), and GCB-DLBCL (p=0.03)(Figure 1d). Moreover, all IHC-positive samples displayed a transcriptional signature of NF-kB pathway activity. The fraction of cases presenting high NF-kB transcriptional activity by GSEA was higher than that defined by immunohistochemistry (>95% ABC-DLBCL and ~47% GCB-DLBCL)(Figure 1e,f). This difference likely reflects the higher sensitivity of GEP-based approaches, but also their inability to discriminate signals deriving from infiltrating reactive cells. Thus, immunohistochemistry may provide a rapid and specific, although relatively less sensitive approach for the identification of constitutively active NF-kB on routine diagnostic material. Both methods revealed that NF-kB signaling is not limited to ABC-DLBCL, but may also be present in a smaller subset of GCB-DLBCL.
Figure 1. The NF-kB pathway is active in ABC-DLBCL and in a smaller fraction of GCB-DLBCL.
a) Immunohistochemical staining of DLBCL biopsies with anti-NFKB1(p105/p50)(top) and anti-NFKB2(p100/p52)(bottom) antibodies. Nuclear localization of NF-kB denotes active signaling, as opposed to the inactive, cytoplasmic complex (200X). b) Prevalence of cases displaying constitutive NF-kB activation in DLBCL subgroups. Color codes indicate nuclear p50, p52 or both. c) Western blot analysis of DLBCL cell lines for processing of p100 to p52 (red, ABC-DLBCL; grey, GCB-DLBCL). The multiple myeloma cell line U266 and the epithelial cell line COS are used as positive and negative controls, respectively. Actin, loading control. d) GSEA enrichment score and distribution of NF-kB target genes along the rank of transcripts differentially expressed in ABC- vs GCB-DLBCL. e) Percentage of samples showing a transcriptional NF-kB signature by GSEA. f) Venn diagram illustrating the overlap between immunohistochemistry-defined and GSEA-defined NF-kB positive cases.
To investigate whether constitutive NF-kB activation in ABC-DLBCL represents a primary pathogenetic event or reflects the intrinsic program of the tumor cell of origin, we screened for mutations the complete coding sequence of 31 NF-kB pathway genes in 14 samples (Table S2). Genes found mutated after filtering for known polymorphisms and synonymous mutations were further analyzed in a validation panel composed of 87 DLBCL (23 ABC, 44 GCB and 20 unclassified/non-GC)(Figure S1).
This strategy identified a total of 48 sequence changes distributed in 6 different genes, including the NF-kB negative regulator TNFAIP3/A2014-16 and the positive regulators CARD118, TNFRSF11A/RANK17, TRAF218,19, TRAF520 and MAP3K7/TAK121 (Table 1 and S3). Mutations were preferentially associated with the ABC-DLBCL phenotype, where 51.3% of the samples analyzed showed alterations in one or more gene, compared to 22.7% GCB-DLBCL (Table 1 and Table S4). In addition, 7/20 (35%) non-GC DLBCL were found mutated. Analysis of paired normal DNA, available from 8 samples, indicated the somatic origin of these events in at least one sample/gene.
Table 1.
NF-kB pathway genes found mutated in DLBCL
| Gene Symbol (alias) | N. of mutated/tested cases (%) |
||
|---|---|---|---|
| ABC-DLBCL | GCB-DLBCL | non-GC/NC DLBCL | |
| TNFAIP3 (A20) | 9/37 (24.3%) | 1/44 (2.3%) | 4/20 (20%) |
| CARD11 | 4/37 (10.8%) | 3/44 (6.8%)* | 2/20 (10%) |
| TNFRSF11A (RANK) | 3/37 (8.1%)^ | 1/44 (2.3%) | 2/20 (10%) |
| TRAF5 | 2/37 (5.4%) | 2/44 (4.5%) | 1/20 (5%) |
| TRAF2 | 1/37 (2.7%) | 4/44 (9.1%)** | 1/20 (5%) |
| MAP3K7 (TAK1) | 2/37 (5.4%) | 0/44 | 0/20 |
| ALL GENES | 19/37 (51.3%) | 10/44 (22.7%)*** | 7/20 (35%) |
One additional cell line was found to carry a mutation in a minority of the population
Two of the 3 mutated samples are cell lines; analysis restricted to exons 4-9, encoding the coiled-coil domain
Three of the 4 mutated samples are cell lines
Four of the 10 mutated samples are cell lines
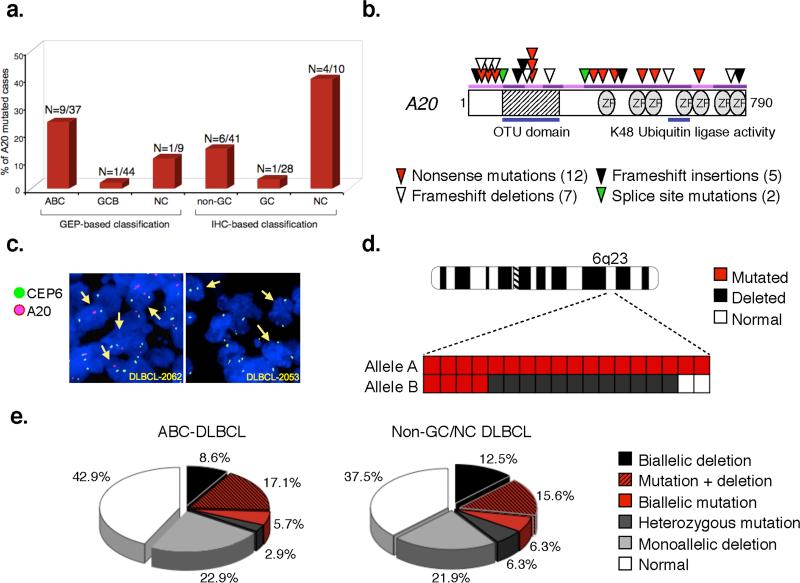
The most commonly affected gene was A20, which encodes for a dual function ubiquitin-modifying enzyme belonging to the ovarian tumor (OTU) domain-containing family of deubiquitinating enzymes and required for termination of NF-kB responses in the classical NF-kB pathway14-16. Notably, the A20 locus is positioned on chromosomal band 6q23.3, a region frequently deleted in aggressive B-cell lymphomas, and suggested to contain a tumor suppressor22,23. We therefore examined this gene in 68 additional DLBCL biopsies, immunohistohemically classified as GC and non-GC based on the Hans algorithm, with minor modifications (see Methods)24. Combined, the two screenings led to the identification of 26 mutational events, distributed in 22 cases and almost exclusively segregating with an ABC/non-GC phenotype (9/37 ABC and 10/51 non-GC/NC DLBCL, vs 2/72 GCB DLBCL) (Figure 2a). Sequence changes included nonsense mutations introducing premature termination codons (N=12); frameshift deletions/insertions (N=7 and 5, respectively); and nucleotide substitutions at consensus splice donor sites (N=2), which were documented by cDNA amplification and sequencing to generate aberrant transcripts that retain intronic sequences and have lost their coding potential (Figure 2b and Table S5). The common consequence of these mutations is the production of severely truncated A20 polypeptides which lack functionally relevant domains (Figure S2) and are either unstable or functionally impaired, as experimentally demonstrated in transient transfection/NF-kB reporter gene assays (Figure S3)14,16.
Figure 2. Mutations and deletions of the A20 gene in ABC-DLBCL.
a) Percentage of A20 mutated cases in various DLBCL subtypes. The exact number over total analyzed is given on top. NC, samples co-expressing CD10 and IRF4 (see Methods). b) Schematic representation of the human A20 protein, with its key functional domains. Color-coded symbols depict distinct alterations leading to A20 inactivation. OTU, ovarian tumor domain; ZF, A-20 zinc-finger domains. c) Dual-color FISH analysis of representative DLBCLs, hybridized with A20-specific probes (red) and a chromosome 6 centromeric probe (green). Arrows point to representative cells displaying hemizygous (left panel) or homozygous (right panel) A20 deletions. d) Chromosome 6 ideogram; the region encompassing A20 on band 6q23 is enlarged in the bottom panel, where columns represent individual cases, and rows correspond to the two alleles. e) Overall frequency of A20 structural alterations (inactivating mutations and deletions, combined).
In 4 samples, each displaying two mutational events, sequencing analysis of A20 transcripts after cDNA amplification and cloning demonstrated that the mutations were located on separate alleles, leading to biallelic gene inactivation. Moreover, FISH analysis using specific probes and/or direct sequencing revealed deletion of the second allele in 12/14 mutated cases with available material (Figure 2c-d, Table S5). Homozygous A20 deletions were found in 7 additional cases, one of which harbored a focal deletion (<420Kb) encompassing A20 and OLIG3, a gene not expressed in B-cells (Figure S4 and Table S6), providing strong evidence for A20 being the target of the lesion. In all samples, loss of the signal accounted for 90% of the tumor cell population, consistent with a clonally represented event (Table S7). Thus, 32% of ABC-DLBCL and ~34% of non-GC/NC DLBCL have lost both copies of the A20 gene due to the presence of inactivating mutations and/or deletions (Figure 2e). Interestingly, monoallelic deletions were also observed in 23% ABC-DLBCL and 22% non-GC DLBCL. Since expression of the wild-type allele was still detected in the 3 cell lines investigated, these data may suggest haplo-insufficiency or the involvement of a second gene in the context of larger 6q chromosomal deletions, frequently observed in aggressive lymphomas22. Collectively, these findings indicate that A20 is frequently inactivated in DLBCL by a two-hit mechanism typical of tumor suppressor genes.
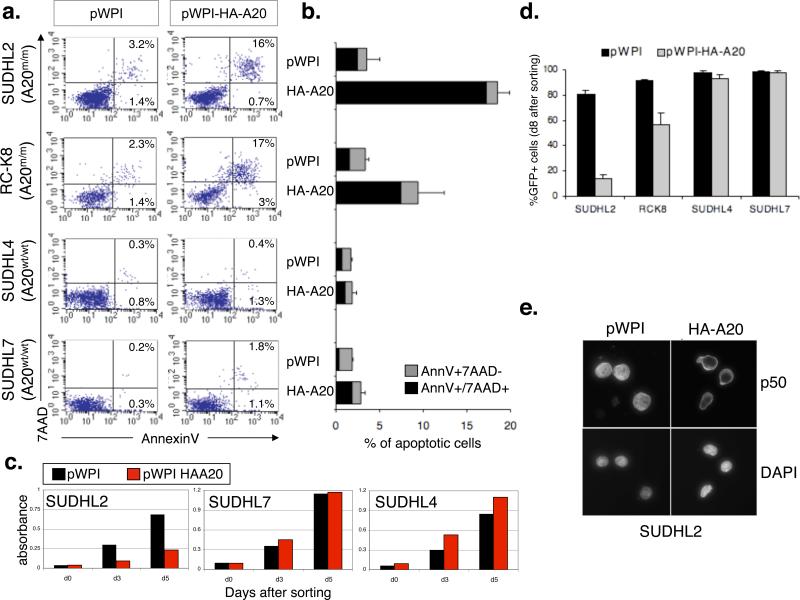
To directly test the role of A20 in cell transformation, we used lentiviral expression vectors and reintroduced A20 in two cell lines (SUDHL2 and RC-K8) carrying biallelic A20 gene inactivation. As shown in Figure 3a-c, A20 reconstitution induced apoptosis and cell growth arrest in the A20-null cell lines, but not in two control lines carrying an intact A20 locus and lacking constitutive NF-kB activity. Consistently, FACS analysis of GFP expression documented the progressive disappearance of the A20-positive population (identified by GFP) in SUDHL2HA-A20 and RC-K8HA-A20, as opposed to SUDHL2WPI and RC-K8WPI, where >90% of the population was GFP+ 8 days after sorting (Figure 3d). Notably, the majority of A20-reconstituted cells showed complete cytoplasmic relocation of p50 by immunofluorescence staining (Figure 3e), indicating an A20-dependent block in NF-kB signaling and consistent with its well-established role in termination of NF-kB responses in vitro and in vivo14-16. Together, these findings strongly suggest a tumor suppressor role for A20, whose loss may contribute to DLBCL pathogenesis by causing supra-physiological activation of NF-kB which, in turn, has oncogenic properties via inhibiting apoptosis and promoting cell proliferation25.
Figure 3. Reconstitution of A20-null cell lines causes apoptosis and cell growth arrest via inhibition of NF-kB.
a, Flow cytometric analysis of AnnexinV-PE/7AAD staining in A20-null and wild-type cell lines, transduced with the indicated vectors; numbers indicate the percentage of single and double-positive cells (lower and upper right quadrants). The results of 3 independent experiments are summarized in panel b (mean ± SD). c, Reduced viability in SUDHL2 cells complemented with A20 expression-vectors (red bars), as compared to vector alone (black bars)(mean±SD, n=3). No effect was observed in two control cell lines. d, Analysis of GFP expression after transduction with pWPI and pWPI-HA-A20 documents the disappearance of the GFP+ population in SUDHL2HA-A20 and RC-K8HA-A20, but not in SUDHL2pWPI and RC-K8pWPI or in A20 wild-type cell lines (mean ± SD, n=2). e) Immunofluorescence analysis of p105/p50 in SUDHL2 cells transduced with pWPI and pWPI-HA-A20 (top). Nuclei are identified by DAPI (bottom).
Less commonly, missense mutations were found in positive regulators of the NF-kB pathway, namely the scaffolding proteins CARD11 (11%), TRAF2 (3%) and TRAF5 (5%), which mediate NF-kB activation via oligomerization and activation of the IKK kinase; the TAK1 serine-threonine kinase (5%), which directly phosphorylates IKK 21,26; and the cell-surface receptor RANK (8.1%), involved in classical NF-kB responses (Table 1 and Table S8). Notably, SNP-array data showed amplification of the regions harboring these genes in 41 cases, suggesting their possible dominant role in activating NF-kB (not shown). To investigate the functional significance of these mutations, we examined their ability to activate a luciferase reporter vector driven by two NF-kB responsive elements in transient transfection assays. In agreement with a recent study3, CARD11 mutations potentiate its NF-kB transactivation activity, in the absence of further stimuli (Figure S6a,b). Significantly enhanced NF-kB activity was also observed upon transfection of the ABC-DLBCL-derived TRAF2-P186R mutant (Figure S6c). When expressed in the DLBCL cell line SUDHL6, which lacks constitutive NF-kB activity, this mutant was sufficient to induce nuclear p50 translocation in most cells, documenting its ability to stimulate this pathway in vivo (Figure S6d). Conversely, no significant differences were associated with 4 GCB-derived TRAF2 mutant alleles and with the mutant TAK1 allele (not shown). Since these mutations were mostly observed in cell lines, their somatic origin could not be verified, leaving open the possibility that they represent non-previously reported polymorphisms. Alternatively, these data may suggest a more subtle effect of the mutations, not detectable by the experimental approach used. While further studies will be required to dissect the significance of these alterations in- vivo, our data show that at least 15/37 (40.5%) ABC-DLBCL (those with A20, CARD11 and TRAF2 alterations) display mutations of proven functional significance in activating NF-kB.
The identification of multiple genetic alterations converging on the same pathway in a sizable fraction of ABC-DLBCL provides a genetic explanation for the presence of constitutive NF-kB activity in this tumor type, suggesting a role for this signaling pathway as a primary pathogenetic event in lymphomagenesis. The most prominent player in this scenario is the known NF-kB negative regulator A20. Notably, structural alterations affecting this gene are also found in Hodgkin lymphoma, PMBL and marginal zone lymphomas27 (Küppers, personal communication; Kato et al, submitted). These findings, together with the evidence of its functional role in modulating NF-kB14-16, identify A20 as a relevant tumor suppressor gene, whose inactivation may contribute to the pathogenesis of several lymphoma subtypes. Since A20 is itself a target of NF-kB and needs to be induced in order to exert its negative feedback effect, additional upstream events are likely required by the tumor cells to activate this signaling cascade and promote selective pressure for A20 inactivation, including engagement of the B-cell receptor by the antigen, CD40-CD40L signaling, and BAFF-BAFFR interaction. However, the observation that, in some cases, multiple genes are simultaneously altered within this signaling pathway, as a combination of positive and negative regulators (for example, A20 and RANK or TAK1) suggests that additional upstream genetic lesions may complement loss of A20.
Constitutive NF-kB activation may promote malignant transformation by providing anti-apoptotic and pro-proliferative signals. Notably, these lesions occur in the same cases displaying structural alterations of BCL6 and BLIMP128,29, which may contribute to lymphomagenesis by suppressing genotoxic responses (BCL6)30 and/or preventing terminal B-cell differentiation (BCL6, BLIMP1)29. As such, these findings provide the rationale and the assays for the identification of DLBCL patients potentially benefiting from targeted anti-NF-kB therapeutic approaches.
METHODS SUMMARY
Characterization of NF-kB activity
The presence of active NF-kB complexes in DLBCL cell lines and primary biopsies was analyzed by IHC/IF analysis of paraffin-embedded tissue sections and cytospin preparations using anti-p105/p50 and anti-p100/p52 antibodies, and by GSEA of NF-kB target genes on Affymetrix U133Plus_2 gene expression profile data.
Mutation analysis
The complete coding sequences and exon/intron junctions of 31 NF-kB genes were analyzed by PCR amplification and direct sequencing of genomic DNA as described29. Mutations were confirmed by sequencing of both strands on independent PCR products, while previously reported polymorphisms, changes present in matched normal DNA and silent mutations were filtered from the analysis.
Fluorescence in situ hybridization (FISH)
FISH analysis was performed on tissue microarrays using two specific BAC probes spanning the A20 gene and a centromeric probe for chromosome 629.
Lentiviral transduction of A20-expression constructs
For the reconstitution assay, DLBCL cell lines were transduced with lentiviral vectors expressing GFP alone (pWPI) or wild-type human A20 linked to IRES-GFP (pWPI-HA-A20), and analyzed for effects on survival, cell growth and NF-kB activity. Productively transduced (GFP+) cells were purified by cell sorting before use in proliferation assays and immunofluorescence staining of nuclear p50.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Methods
DNA extraction, amplification and sequencing
Genomic DNA was extracted according to standard methods. In 8 cases with available matched non-neoplastic tissue, DNA was also extracted from paraffin-embedded material using the QIAamp DNA mini Kit (QIAGEN, Valencia, CA). Sequences for all annotated exons and flanking introns of the 31 NF-kB pathway genes listed in Table S2 were obtained from the UCSC Human Genome database, using the corresponding mRNA accession number as a reference. The Primer 3 program (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) was used to design oligonucleotides for amplification and sequencing of each coding exon (plus ~50bp of adjacent introns), available upon request. The primers used for analysis of the 6 genes found mutated are reported in Table S9. Purified amplicons were sequenced directly from both strands as described, and compared to the corresponding germline sequences, using the Mutation Surveyor Version 2.41 software package (Soft Genetics LLC)29. Synonymous mutations, changes due to previously reported polymorphisms (Human dbSNP Database at NCBI, Build 129, and Ensembl Database) and changes present in normal DNA from the same patient, when available, were excluded. Somatic mutations were confirmed on independent PCR products. In cases displaying more than one event within a single gene (A20, CARD11 and TRAF5), the allelic distribution of the mutations was determined by cloning and sequencing full-length PCR products obtained from cDNA (N= 10 clones each)31.
Tissue microarrays, immunohistochemistry and immunofluorescence staining
The construction of the DLBCL tissue microarray was performed according to standard procedures and the protocols for immunohistochemical and immunofluorescence staining are described in Ref. 13. Samples were classified as GC or non-GC types based on expression of CD10, BCL6 and IRF4 according to Hans et al.24, except that the rare CD10+ cases co-expressing IRF4 were designated as non-classified (NC), since IRF4 is a known marker of B-cell activation, and is normally absent in BCL6+ GC centroblasts32. The percentage and staining intensity of neoplastic B-cells were independently scored by two pathologists (A.Ch. and G.B.), using a cut-off of 30% positive cells. Cases were considered to be positive for NF-kB activity when ≥30% of tumor cells showed nuclear NF-kB localization. The antibodies used were rabbit monoclonal anti NF-kB1 p105/p50 and NF-kB2 p100/p52 (18D10) (Cell Signaling Technology).
RNA extraction, generation of gene expression profiles and DLBCL classification
The protocols for RNA extraction, cRNA labeling and hybridazion to Affymetrix GeneChip U133Plus_2 microarrays are described in detail in Ref. 13. Gene expression data were normalized by the MAS 5.0 software, followed by log2 transformation. The DLBCL primary biopsies were classified into GCB (N=38), ABC (N=30) and unclassified (N=9) as previously described6, using a linear predictive score and 22 of the 27 original lymphochip predictor genes which were represented in the U133Plus_2 array and showed the best t-statistics. Cases displaying inconsistencies between COO-classification, unsupervised hierarchical clustering analysis and immunhistochemistry-based classification were considered as unclassified or excluded from further GEP-based analyses.
Gene Set Enrichment Analysis (GSEA)
Enrichment analysis of NF-kB target genes was performed as previously described33 using the genes listed in Table S1 and gene expression profiles from the DLBCL biopsies (GSEA v2.0 at www.broad.mit.edu/gsea).. The NF-kB target gene set was generated by combining previously reported target genes identified in GEP studies of B-cells, and included genes that were specifically downregulated after genetic (induction of NF-kB super-repressor; CARD11 shRNA) or pharmacologic (IKK-inhibitors) manipulation of NF-kB in representative ABC-DLBCL and Hodgkin lymphoma cell lines. GSEA was also used to assess whether individual DLBCL samples expressed a transcriptional signature of NF-kB activation. To this end, the expression of each gene on the U133Plus_2 microarray was first converted into z-score using 10 samples of purified normal GC B-cells as a baseline. Genes were then ranked by their z-score from the most positive to the most negative value, and the 120 genes of the NF-kB gene set were intersected with the ordered list to compute GSEA Enrichment Scores (ES). The algorithm was set to implement weighted scoring scheme and the ES significance was assessed by 100000 permutation tests. Samples attaining significant p-value (p<0.05, Bonferroni corrected) were designated as samples with activated NF-kB.
Fluorescence in situ hybridization (FISH)
Two PAC clones (RP11-703G8 and RP1-702P5) spanning the A20 gene were obtained from BACPAC Resources at http://bacpac.chori.org. DNA was labeled by nick-translation using spectrum orange dUTP fluorochrome (Vysis). A Spectrum green-labeled centromeric probe (Vysis Inc., Downers Grove, IL) was used to enumerate chromosome 6. Paraffin-embedded tissue sections from TMAs were baked overnight at 60oC and processed using a paraffin pretreatment kit (Vysis Inc., Downers Grove, IL). FISH was performed on 4’,6’-diamidino-2-phenylindole (DAPI)-stained slides by standard methods, and hybridization signals were scored on at least 500 interphase nuclei/core (i.e., five representative areas with at least 100 nuclei each). Slides were evaluated for probe signal intensity and signal to background ratio. As control, multiple sections from normal tonsils were included in each TMA. Normal variation corresponded to 9.7+/-4.6% of the nucleated cells for loss of 6q23 signal, and 33.5+/-12.5 for monosomy 6. Cases were diagnosed as positive when the fraction of cells showing an abnormal pattern was above the mean +2SD (+1SD for monosomies). The percentage of tumor cells in each core was estimated by histologic analysis of serial TMA sections.
Lentiviral infections
The replication deficient lentiviral expression construct pWPI-HA-A20 was generated by subcloning the full-length A20 cDNA sequences into the PmeI restriction site of pWPI, in front of IRES-GFP. Viral supernatants were obtained by co-transfecting 293T cells with the lentiviral expression vectors and vectors expressing the helper virus 8.9 and the VSV-G envelope glycoprotein34,35. Conditioned medium was harvested over 48-62 hours and used directly to infect the indicated cell lines according to standard methods. Transduction efficiencies were determined by FACS analysis of GFP expression after 48-72 hours. For cell proliferation assays, western blot analysis of exogenous A20 expression, and immunofluorescence staining of nuclear p105/p50, productively infected cells were sorted by flow cytometry on a BD FACSAria Cell Sorter, based on GFP expression.
Apoptosis and proliferation assays
The effect of A20 expression on cell survival was measured 48 and 72 hrs after infection by flow cytometric analysis of AnnexinV-PE and 7-amino-actimomycin D (7AAD; BD Pharmingen Biosciences) stained cells, gating on the GFP+ population. Data were acquired on a FACSCalibur (Becton Dickinson) and analyzed with the CELLQuest software. Cell proliferation was monitored on sorted GFP+ cells using the MTT reagent (Roche) according to the manufacturer's instructions. The fraction of live GFP+ cells was also measured over time by FACS analysis, and compared to the initial GFP+ population (determined 2 days after lentiviral transduction) on at least three independent experiments.
Supplementary Material
Acknowledgements
We thank P. Smith, P. Chadwick and the Molecular Pathology Facility of the Herbert Irving Comprehensive Cancer Center (HICCC) at Columbia University Medical Center for histology service; VVV Murty and the HICCC Molecular Cytogenetics Service for assistance on the FISH analysis; the HICCC Flow Cytometry Facility for fluorescence-activated cell sorting; V. Miljkovic and J. Pack for help with the Affymetrix gene expression hybridization; U. Klein and D. Dominguez-Sola for suggestions; L. Menard for help with the mutation analysis, and G. Inghirami for the pWPI lentiviral vector. Automated DNA sequencing was performed at Genewiz.Inc. L.P. is on leave from the Institute of Hematology, University of Perugia Medical School, Perugia, Italy. This work was supported by NIH grants P01 CA92625-07 (R.D-F), NIAID R01AI066116, the National Centers for Biomedical Computing NIH Roadmap initiative U54CA121852 (A.Califano), and a Leukemia & Lymphoma Society SCOR grant (R.D.-F). L.P. would like to dedicate this work to the memory of Enrico Pasqualucci.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author contributions: L.P. and R.D.F. designed the study. M.C., A.G. and Q.S. performed experiments; L.P. performed the A20 functional assays; W-K. Lim and A.Califano developed tools for GEP analysis; S.V.N. performed the FISH analysis; A.C. and G.B. analyzed all immunohistochemistry data; A.C., G.B., F.B. and M.P. provided DLBCL samples; M.B., F.B, and M.S. performed SNP array analysis; L.P. designed experiments, coordinated the study, analyzed data and wrote the manuscript, which was commented by all authors.
Author Information: Reprints and permissions information is available at www.nature.com/reprints. Correspondence and requests for materials should be addressed to: lp171@columbia.edu
Accession numbers. The expression data and the 250K SNP data reported in this paper have been deposited in the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) database (Series Accession Number GSE12195 and GSE15127).
References
- 1.Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–11. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 2.Davis RE, Brown KD, Siebenlist U, Staudt LM. Constitutive nuclear factor kappaB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J Exp Med. 2001;194:1861–74. doi: 10.1084/jem.194.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lenz G, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319:1676–9. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- 4.Monti S, et al. Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood. 2005;105:1851–61. doi: 10.1182/blood-2004-07-2947. [DOI] [PubMed] [Google Scholar]
- 5.Savage KJ, et al. The molecular signature of mediastinal large B-cell lymphoma differs from that of other diffuse large B-cell lymphomas and shares features with classical Hodgkin lymphoma. Blood. 2003;102:3871–9. doi: 10.1182/blood-2003-06-1841. [DOI] [PubMed] [Google Scholar]
- 6.Wright G, et al. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci U S A. 2003;100:9991–6. doi: 10.1073/pnas.1732008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ngo VN, et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–10. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- 8.Rawlings DJ, Sommer K, Moreno-Garcia ME. The CARMA1 signalosome links the signalling machinery of adaptive and innate immunity in lymphocytes. Nat Rev Immunol. 2006;6:799–812. doi: 10.1038/nri1944. [DOI] [PubMed] [Google Scholar]
- 9.Li Q, Verma IM. NF-kappaB regulation in the immune system. Nat Rev Immunol. 2002;2:725–34. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 10.Hoffmann A, Baltimore D. Circuitry of nuclear factor kappaB signaling. Immunol Rev. 2006;210:171–86. doi: 10.1111/j.0105-2896.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- 11.Pomerantz JL, Baltimore D. Two pathways to NF-kappaB. Mol Cell. 2002;10:693–5. doi: 10.1016/s1097-2765(02)00697-4. [DOI] [PubMed] [Google Scholar]
- 12.Coope HJ, et al. CD40 regulates the processing of NF-kappaB2 p100 to p52. Embo J. 2002;21:5375–85. doi: 10.1093/emboj/cdf542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basso K, et al. Tracking CD40 signaling during germinal center development. Blood. 2004;104:4088–96. doi: 10.1182/blood-2003-12-4291. [DOI] [PubMed] [Google Scholar]
- 14.Boone DL, et al. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5:1052–60. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 15.Lee EG, et al. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 2000;289:2350–4. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wertz IE, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–9. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 17.Anderson DM, et al. A homologue of the TNF receptor and its ligand enhance T-cell growth and dendritic-cell function. Nature. 1997;390:175–9. doi: 10.1038/36593. [DOI] [PubMed] [Google Scholar]
- 18.Rothe M, Sarma V, Dixit VM, Goeddel DV. TRAF2-mediated activation of NF-kappa B by TNF receptor 2 and CD40. Science. 1995;269:1424–7. doi: 10.1126/science.7544915. [DOI] [PubMed] [Google Scholar]
- 19.Yeh WC, et al. Early lethality, functional NF-kappaB activation, and increased sensitivity to TNF-induced cell death in TRAF2-deficient mice. Immunity. 1997;7:715–25. doi: 10.1016/s1074-7613(00)80391-x. [DOI] [PubMed] [Google Scholar]
- 20.Rothe M, Wong SC, Henzel WJ, Goeddel DV. A novel family of putative signal transducers associated with the cytoplasmic domain of the 75 kDa tumor necrosis factor receptor. Cell. 1994;78:681–92. doi: 10.1016/0092-8674(94)90532-0. [DOI] [PubMed] [Google Scholar]
- 21.Yamaguchi K, et al. Identification of a member of the MAPKKK family as a potential mediator of TGF-beta signal transduction. Science. 1995;270:2008–11. doi: 10.1126/science.270.5244.2008. [DOI] [PubMed] [Google Scholar]
- 22.Gaidano G, et al. Deletions involving two distinct regions of 6q in B-cell non-Hodgkin lymphoma. Blood. 1992;80:1781–7. [PubMed] [Google Scholar]
- 23.Honma K, et al. TNFAIP3 is the target gene of chromosome band 6q23.3-q24.1 loss in ocular adnexal marginal zone B cell lymphoma. Genes Chromosomes Cancer. 2008;47:1–7. doi: 10.1002/gcc.20499. [DOI] [PubMed] [Google Scholar]
- 24.Hans CP, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–82. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 25.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–6. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 26.Wang C, et al. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–51. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 27.Novak U, et al. The NF-kB negative regulator A20 (TNFAIP3) is commonly inactivated by somatic mutations and genomic deletions in marginal zone B-cell lymphomas. Blood. 2009 doi: 10.1182/blood-2008-08-174110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saito M, et al. A signaling pathway mediating downregulation of BCL6 in germinal center B cells is blocked by BCL6 gene alterations in B cell lymphoma. Cancer Cell. 2007;12:280–92. doi: 10.1016/j.ccr.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 29.Pasqualucci L, et al. Inactivation of the PRDM1/BLIMP1 gene in diffuse large B cell lymphoma. J Exp Med. 2006;203:311–7. doi: 10.1084/jem.20052204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432:635–9. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- 31.Pasqualucci L, et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341–6. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- 32.Falini B, et al. A monoclonal antibody (MUM1p) detects expression of the MUM1/IRF4 protein in a subset of germinal center B cells, plasma cells, and activated T cells. Blood. 2000;95:2084–92. [PubMed] [Google Scholar]
- 33.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–72. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 35.Naldini L, et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–7. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.