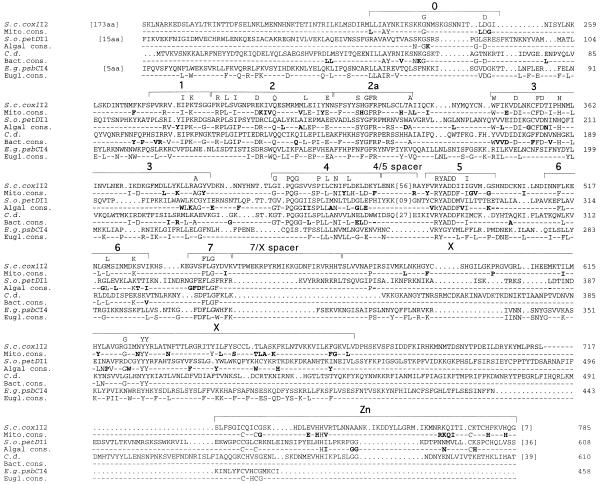
Figure 2.
Alignment of RT amino acid sequences. One example sequence is shown for each of the major phylogenetic groupings (mitochondrial, algal, bacterial and euglenoid; see Fig. 3). Below each example sequence is the consensus sequence for that group, defined as the positions conserved in at least 75% of the members. The overall consensus is presented above the alignment, and is defined as residues common to the mitochondrial, algal and bacterial consensus sequences. Domains 0–7, X and Zn are marked as described in the text. The locations of spacers between RT domains 4 and 5 and between domains 7 and X are indicated (see text and Fig. 3). Residues marked in bold are consensus sequence positions specific to one group. In cases of closely related sequences (e.g. plant nad1I4 introns), only one example was included in the consensus sequence calculation. Sequences used in the calculation were: mitochondrial group (Z.m.nad1I4, M.p.cox1I1, M.p.cob1I3, M.p.atpAI1, M.p.atpAI2, S.c.cox1I2, S.c.cox1I1, M.p.atp9I1, S.p.cox2I1, S.p.cox1I1, A.m.cox1I3, P.a.cox1I4, M.p.SSUI1, P.a.cox1I1, S.a.1, L.l., S.p.cob1I1, V.i.cob1I1, P.li.cox1I3, P.a.ND5I4, M.p.ORF732, M.p.cox1I2, P.li.cox1I2, P.li.cox1I1, N.c.cox1I1 and M.p.cox2I2); algal group (P.p.LSUI1, C.s., B.m.rbcLI1, B.a.-07, S.o.petDI1, P.li.LSUI1, P.li.LSUI2, P.s. and E.c.-0157); bacterial group (S.f., S.p., P.a., B.m., C.d., B.a.-23, S.me., B.h. and E.c.B); and euglenoid group (E.v.psbCI4, A.l.ORF456, E.g.psbCI4, E.d.psbCI4, L.b.psbCI4, E.m.psbCI4 and P.s.cpn60I1).