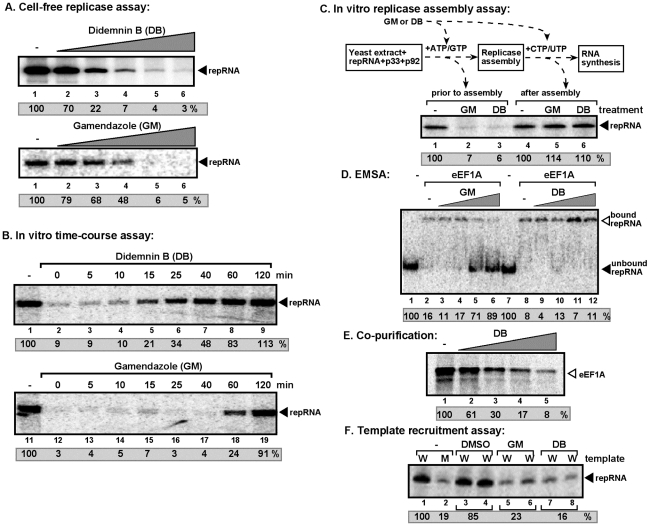
Figure 4. Inhibition of TBSV repRNA replication by Didemnin B and Gamendazole in a cell-free TBSV replicase assay.
(A) The cell-free TBSV replicase assay was performed as described in Fig. 2. DB and GM were added in the following amounts: 0, 50, 100, 150, 200, 250 µM for DB and 0, 5, 25, 50, 100, and 200 µM for GM, respectively. The replicase activity in the samples containing the DMSO solvent instead of DB or GM was chosen as 100%. (B) Time course analysis was performed in a cell-free TBSV replicase assay as described in Fig. 2. DB (150 µM) and GM (100 µM) were added at various time points and the replicase assay was stopped after 3 hours for each treatment, followed by RNA analysis in a denaturing PAGE gel. The replicase activity in the samples containing DMSO added at the 0 time point was chosen as 100%. (C) A step-wise approach was used to separate the possible effect of DB and GM during either the assembly of the TBSV replicase or RNA synthesis steps. In step 1, the purified recombinant TBSV p33, p92pol and (+)repRNA were added to the whole cell extract in the presence of ATP and GTP, which only supports the assembly of the TBSV replicase, but prevents RNA synthesis. This was followed by removal of the extra amount of p33, p92pol and repRNA, which were not bound to the membranes of cell-free extract, and then by the standard replicase assay in a buffer containing 32P-UTP and ATP, CTP and GTP (step “RNA synthesis”). The denaturing PAGE analysis of the 32P-labeled repRNA products obtained is shown. Note that DB (150 µM) and GM (100 µM) were added to the assay either at the beginning (prior to replicase assembly) or after the replicase assembly. See further details in panel B. (D) The effect of DB and GM on binding between the purified eEF1A and 32P-labeled template RNA (SL1/SL2/SL3) based on EMSA. The bound and unbound RNAs are pointed at by arrowheads. GM and DB were applied in the following amounts: 0, 5, 50, 250, and 1000 µM. Note that the amount of unbound RNA in the absence of eEF1A (lane 1) was chosen as 100%. (E) The inhibitory effect of DB on co-purification of eEF1A with the viral repRNA. WT 35S-labeled eEF1A was produced in a translation assay using rabbit reticulocyte lysate, followed by incubation with biotin-labeled DI-72(+) repRNA in the presence of 0, 50, 150, 500 and 1000 µM DB. Then the repRNA was captured with streptavidin-coated magnetic beads, followed by elution of the co-purified proteins from the beads. SDS-PAGE analysis shows the amount of co-purified 35S-labeled eEF1A. (F) The inhibitory effect of DB and GM on the template recruitment step in vitro. Purified recombinant p33/p92 and 32P-labeled DI-72 (+)repRNA (indicated as W, lanes 1 and 3–8) or C99-G mutant (+)repRNA (indicated as M, lane 2) were added to a whole cell extract (CFE) in the presence of DMSO (control), 100 µM GM or 150 µM DB, followed by centrifugation/washing to remove the 32P-labeled repRNA that is not bound to the membrane. Then the membrane-bound RNA was analyzed in a denaturing PAGE gel. Note that the recruitment deficient C99-G mutant repRNA bound to the membrane nonspecifically (∼20% level).