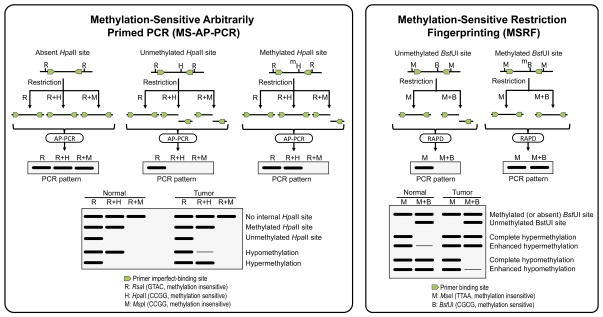
Figure 4.
Methylation-Sensitive Arbitrarily-Primed PCR (left) and Methylation-Sensitive Restriction Fingerprinting (right) rely on the amplification of DNA libraries generated by methylation-sensitive and methylation-insensitive restriction enzymes. MS-AP-PCR libraries are generated by digestion with RsaI (GTAC) alone or in combination with HpaII (CCGG, methylation-sensitive) or MspI (CCGG, methylation-insensitive). After digestion, PCR amplification of the DNA fragments libraries is performed with 20-mer primers following the principle of AP-PCR. MSRF libraries are generated by treatment of the genomic DNA with MseI (TTAA, methylation-insensitive) alone or in combination with BstUI (CGCG, methylation-sensitive), and the amplification is performed with a pair of shorter primers, following the RAPD protocol. In both techniques, changes in the intensity of the fingerprinting bands reflect changes in methylation of the methylation-sensitive enzyme recognition sites.