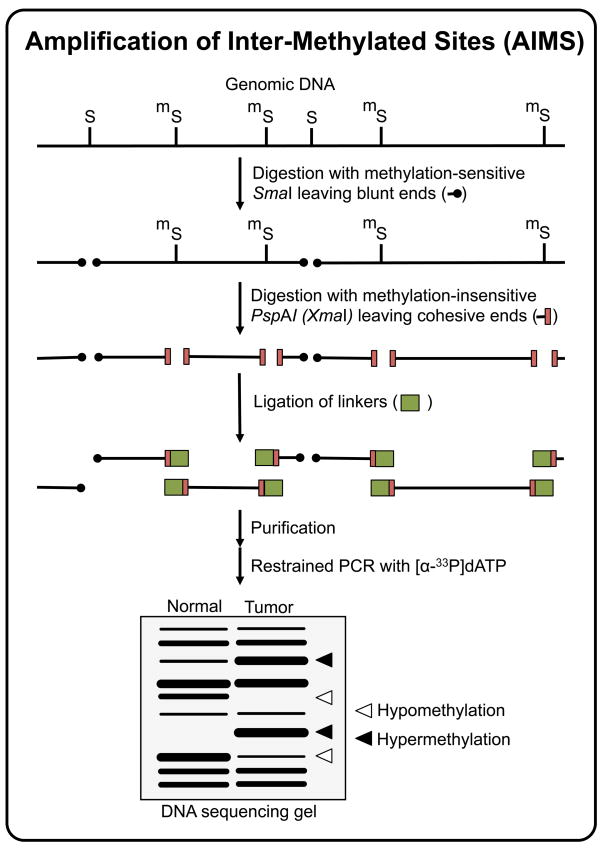
Figure 5.
Amplification of Intermethylated Sites (AIMS) technique. Non-methylated SmaI recognition sites (CCCGGG) are first digested using the methylation-sensitive SmaI restriction endonuclease, leaving blunt ends. A second digestion is then performed using the methylation-insensitive isoschizomer XmaI, leaving 5’ protruding ends to which compatible adaptors are ligated. The DNA fragments in the range of ~200 to ~2000 bp and flanked by two ligated adaptors are subsequently amplified using primers that anneal to the adaptor sequence plus the restriction sequence and, to limit the number of amplified fragments, 1 or more arbitrarily chosen additional nucleotides. The increase of intensity of a particular AIMS fingerprint band reflects the hypermethylation of the flanking SmaI sites in the original sequence, while a decrease of intensity indicates hypomethylation.