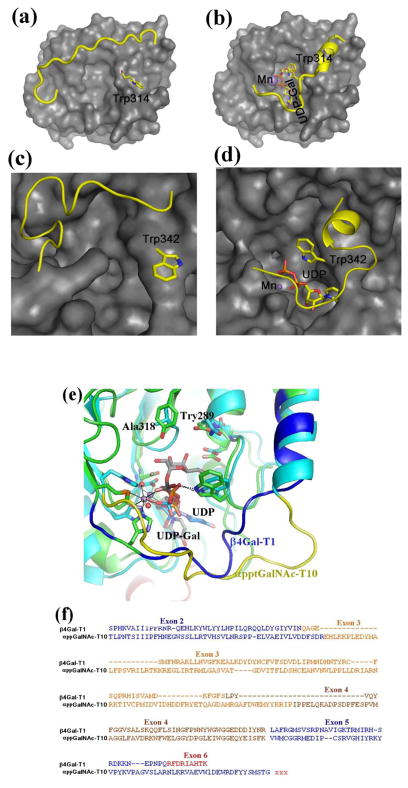
Figure 3.
The crystal structure of the bovine β4Gal-T1 in the open (a) and closed (b) conformation. Upon the binding of manganese and UDP-Gal, the enzyme undergoes conformational changes involving a short and a long flexible loop. In the short loop, the side chain of the Trp314 residue moves from outside to inside the catalytic pocket to bind to the bound UDP-Gal molecule, while the long loop moves over the bound UDP-GalNAc molecule to cover it. The crystal structure of the human αppt-GalNAc-T2 is also found in an open (c) and a closed (d) conformation. Similar to β4Gal-T1, upon the binding of manganese and UDP-GalNAc molecule, this enzyme also undergoes conformational changes involving two flexible loops. Also, in the short loop, the Trp342 residue moves from outside to inside the catalytic pocket to bind to the bound UDP-GalNAc molecule, while the long flexible loop covers the bound UDP-GalNAc molecule.. (e) The superposition of the catalytic domains of bovine β4Gal-T1•Mn2+•UDP-Gal complex with the αppGalNAc-T10•Mn2+•UDP-GalNAc complex and (f) the corresponding protein sequence comparison. The Trp residue in the short flexible loops of these structures show good agreement, and the long flexible loop of β4Gal-T1 and αpptGalNAc-T10 are shown in blue and yellow, respectively. The Tyr289 residue in the bovine β4Gal-T that determines the donor sugar specificity is naturally present as Ala318 residue in αppGalNAc-T10 in order to accommodate the N-acetyl moiety of the donor sugar GalNAc, and it is conserved in all αppGalNAc-T enzymes. In the protein-protein sequence comparison (f), the amino acids are colored, based on their exons. Since the acceptor substrate for β4GalNAc-T1 is a sugar residue, while it is a linear peptide with a Thr/Ser amino acid for the αppGalNAc-T10, these enzymes are expected to have different acceptor binding sites. The additional amino acids in the exons 3 and 4 of αppGalNAc-T10 are found as a part of the acceptor substrate binding site in the αppGalNAc-T10 crystal structure.