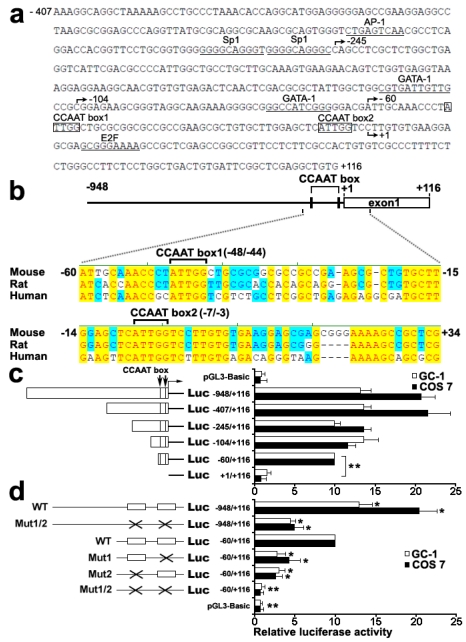
Figure 1.
Identification and function characterization of the mouse Dmrt7 promoter. a), Nucleotide sequence of the promoter region (from -407 to +116) of mouse Dmrt7. The putative binding sites for transcriptional factors are underlined. The CCAAT boxes are boxed. The numbering of the nucleotides starts at the first nucleotide of the cDNA (+1). b), Sequence alignment of 1 kb upstream of the first exon of Dmrt7 from mouse, rat and human. Two highly conserved CCAAT boxes are indicated. Conserved nucleotides among all three species are denoted by red letters and are shaded in yellow. The positions of the nucleotides in the mouse Dmrt7 gene are indicated. +1 represents the first nucleotide of the cDNA. c), Deletion analysis of the mouse Dmrt7 promoter using the luciferase assay. GC-1 or COS 7 cells were transiently transfected with 0.4 µg of the deletion constructs together with 10 ng Renilla luciferase plasmid, pRT-TK, which served as an inner control for transfection efficiency. The relative activities to the -60/+116 construct of a series of deletion mutants was determined by the luciferase assay. The results are the mean ± S.D. of three independent experiments. *, P < 0.05; **, P < 0.001. d), Mutation analysis in the CCAAT boxes of the Dmrt7 promoter by the luciferase assay. The -948/+116 and -60/+116 constructs were used as a template to generate point mutants. GC-1 or COS 7 cells were transiently transfected with 0.4 µg of these constructs together with 10 ng Renilla luciferase plasmid, pRT-TK. The relative activities of the -60/+116 construct of these mutants was determined by the luciferase assay. The results are the mean ± S.D. of three independent experiments. *, P < 0.05; **, P < 0.001 with compared to the -60/+116 wild-type construct. The box indicates the intact CCAAT box. The fork indicates the corresponding mutations.