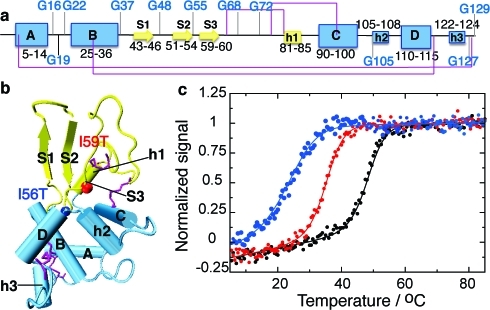
Figure 1.
Structure of human lysozyme indicating the location of destabilizing mutations. (a) Representation of the secondary structure of the native state of WT human lysozyme, indicating glycine residues (using DSSP(17) on pdb entry 1LZ1(18)). Strands are represented by arrows, α-helices (A to D) by large boxes, and 310 helices (h1 to h3) by small boxes. The disulfide bridges C6−C128, C30−C116, C65−C81, and C77−C95 are shown in purple. The α and β domains are shown in blue and yellow, respectively. (b) Tertiary structure of WT human lysozyme with the same color coding as in panel a. All structures are drawn using vmd(19) with the secondary structure defined by DSSP.(17) Vmd only represents strands comprising three residues and more; therefore, strand 3 (residues 59−60) is not shown. The colored spheres represent residues 56 (blue) and 59 (red), sites of the mutations I56T and I59T studied herein. (c) Normalized signal of the near-UV circular dichroism thermal unfolding traces obtained for the WT (black), I59T (red) and I56T (blue) variants, at pH 1.2.