Figure 2.
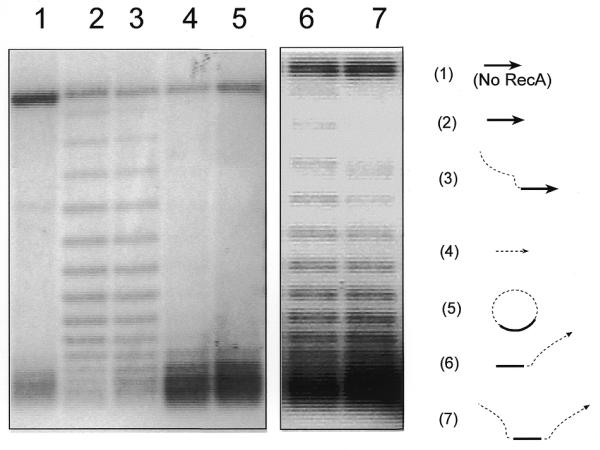
A circular RecA–ssDNA filament cannot unwind its supercoiled target DNA. Using the assay described in Figure 1A, shown here is an agarose gel with 2 mg/ml chloroquine. The diagrams to the right of the gel indicate the ssDNA substrates used in each reaction; the target duplex DNA used in all reactions was the 3 kb supercoiled pBluescript plasmid. The solid line indicates the homologous region (120 nt) and the dashed line indicates the heterologous region (400 nt). Lane 5 shows a reaction in which circular RecA–ssDNA filament (C-520, 520 nt long with 120 nt homologous to the target duplex DNA) was used as the searching entity. Lane 3 shows the same reaction but linear 520mer ssDNA (with the 3′-end 120 nt homologous to the target duplex) was used as the searching entity. Lane 2 is the reaction in which a 120mer homologous ssDNA (H-120) was used. Lanes 1 and 4 are control reactions either without RecA protein or with heterologous 124mer ssDNA (Het-124). Lanes 6 and 7 show two reactions with linear 520mer ssDNA with 120 nt homologous to the target duplex located at the 5′-end (lane 6) or at the center (lane 7).
