Figure 3.
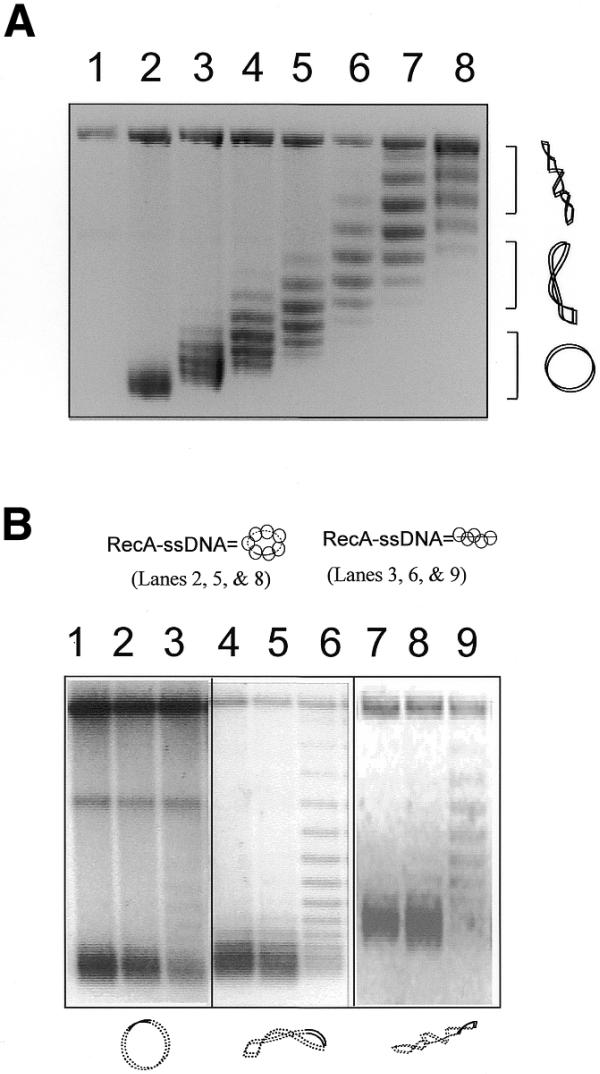
Unwinding of target DNAs with different degrees of supercoiling by a linear RecA–ssDNA filament. (A) An agarose gel with chloroquine (2 mg/ml) shows the covalently closed circular duplex DNA (pBluescript, 3 kb with 120 bp homologous to the ssDNA used) with high to zero supercoiling that was used in the reactions shown in (B). In the chloroquine gel the relaxed duplex DNA will become positively supercoiled. Therefore, it runs faster than the negatively supercoiled DNA that has become relaxed under these conditions. Lane 1 is a nicked circle as the marker for relaxed plasmid. Lanes 2–8 are the covalently closed plasmids (pBluescript, 3 kb) with increasing negative superhelicity. (B) An agarose gel with chloroquine (2 mg/ml) showing unwinding of the circular target DNA caused by either linear or circular homologous RecA–ssDNA. The diagrams at the top and bottom of the gel indicate the RecA–ssDNA and the duplex target DNA used in the reactions; solid lines represent homology between the two molecules. Lanes 1–3 are reactions where relaxed duplex was used as the target. Lanes 4–6 are reactions where medium supercoiled duplexes (superhelical density ∼–0.016) were used as target. Lanes 7–9 are reactions where highly supercoiled duplex (superhelical density ∼–0.033) were used as the target. Lane 3, 6 and 9 show reactions in which a homologous linear 120mer RecA–ssDNA filament (H-120) was used as the searching molecule. Lanes 1, 4 and 7 are reactions in which 124mer heterologous RecA–ssDNA filament (Het-124) was used as the searching molecule. Lanes 2, 5 and 8 show a reaction in which circular RecA–ssDNA filament (C-520, 520 nt long with 120 nt homologous to the target duplex DNA) was used as the searching molecule.
