Figure 6.
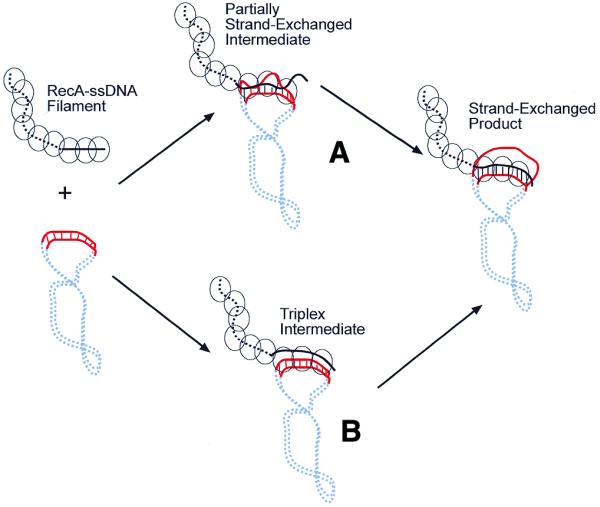
Proposed model for RecA-mediated homology recognition. (A) Homology recognition is likely coupled to local strand exchanges between the two molecules. These locally unwound regions on the target duplex DNA could either be induced through contacts with the RecA–ssDNA filament or simply be the result of thermal fluctuation of the duplex DNA. Thus, the searching RecA–ssDNA filament tests for homology by base pairing with its complementary strand in the target duplex DNA. If homology is found through these short strand exchanges, then branch migration can extend the exchanged region to a greater distance, eventually resulting in full strand exchange. (B) In this model the RecA–ssDNA forms a triplex intermediate with the target duplex DNA prior to strand exchange. The results presented in this paper do not support this model, where in the triplex intermediate the RecA–ssDNA (which has been extended to ∼18 bp/turn) needs to be aligned in register with its target duplex DNA (10 bp/turn) on the homologous region, yet does not unwind the target duplex DNA.
