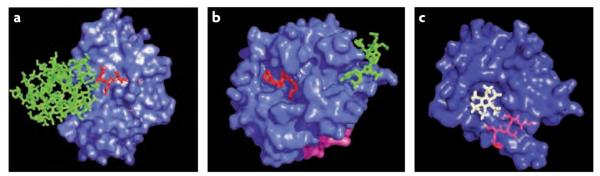
Figure 2. Active site, exosite and allosteric site.
Three proteases of known structure have been chosen to illustrate the components identified in many (but not all) proteases that define their specificity and their control of activity. Surfaces of sentrin-specific protease 2 (SENP2) (Protein Data Bank code 1TGZ125) (a) and thrombin (Protein Data Bank code 1EB1 (REF. 126)) (b) with substrate or inhibitor residues contacting the active site cleft (shown in red sticks) and with substrate or inhibitor residues contacting the exosite (shown in green sticks) are shown. In SENP2, the exosite is composed of a large disperse region that occupies the bulk of the small ubiquitin-like modifier (SUMO) substrate, which is required for directing specificity and enhancing activity76. By contrast, the thrombin exosite consists of a relatively small cationic patch that is required for efficient catalysis of fibrinogen and for binding to the inhibitor hirudin21. The surface of Vibrio cholerae RtxA toxin (Protein Data Bank code 3GCD127) (c) centres on the allosteric natural small-molecule activator inositol hexakisphosphate (shown in white), with the active site bound peptide (shown in red) underlying a loop in the translucent surface. In the case of SENP2, the exosite is potentially quite large, complicating small-molecule control. However, in thrombin and RtxA the exosite or the allosteric site is composed of surfaces compatible with small-molecule targeting.