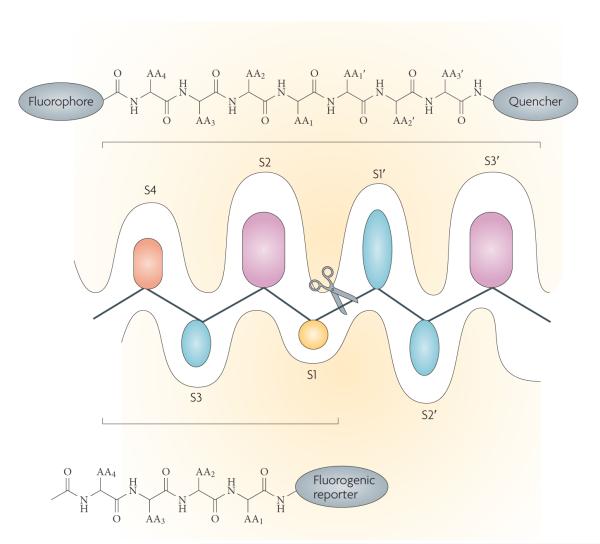
Figure 3. Probing protease active sites.
In standard protease notation, substrates are cleaved between the P1 (amino-terminal) and P1′ (carboxy-terminal) residues, with P1,2,3 … n and P’1,2,3 … n residues increasing towards the N terminus of the protein and towards the C terminus respectively. The corresponding pockets on the protease that accept the substrate side chain are designated S or S’ accordingly128. The schematic representation of the active site cleft of proteases shows the prime S1′-S3′ pockets accommodating specific amino acid side chains identified on the C-terminal side of the scissile bond and unprimed sites S1–S4 identified on the N-terminal side of the scissile bond (indicated by scissors). Shown also are the types of chemical moiety that can be used to investigate protease specificity either as individual substrates or as combinatorial libraries. The application of positional scanning substrate libraries allows exploration of the individual pockets of the protease by synthesis of a set of sublibraries. These generally consist of natural amino acids linked to a fluorogenic reporter, or, for a wider exploration of the catalytic cleft, of peptides that contain fluorophores and a quencher. The cleavage preference for each sublibrary is determined from fluorescence yields generated when the fluorophore is separated from the quencher by cleavage of the peptide bond.