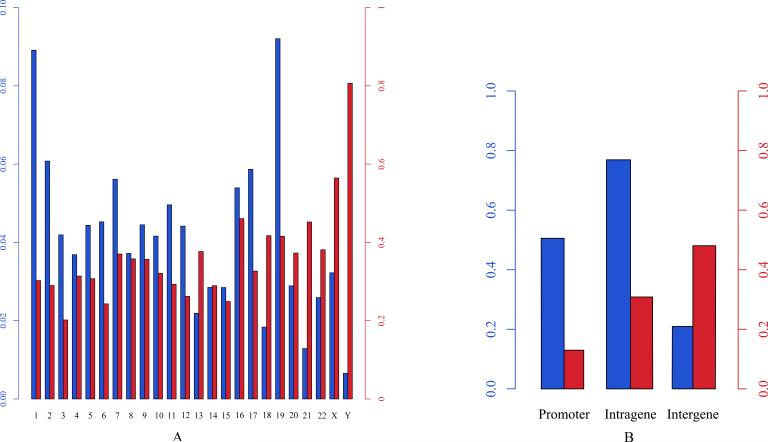
Figure 4. The distribution of CGIs and methylation-prone CGIs.
(A) The distribution of the number of CGIs in chromosomes (blue bar) and the proportion of methylation-prone CGIs in each chromosome (red bar). One can see that there are the most CGIs in chr19 and the least CGIs in chrY, and more than 80% of the CGIs located in chrY are prone to DNA methylation, while 33.16% in autosome are methylation-prone. (B) The distribution of the number of CGIs in promoters, intragenic and intergenic regions (blue bar), and the proportion of methylation-prone CGIs located in promoters, intragenic and intergenic regions (red bar). One can see that less than 13% of the CGIs located in promoter regions are prone to DNA methylation.