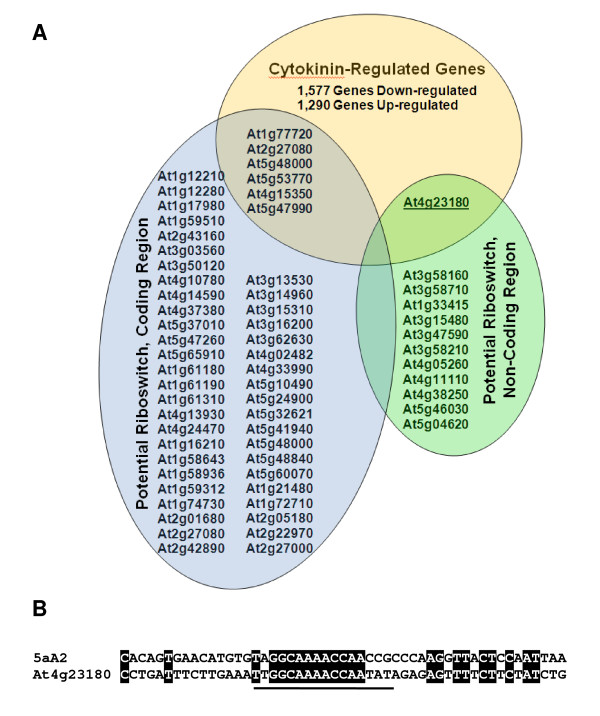
Figure 2.
Candidate cytokinin-binding riboswitch aptamers (CaAps). Three data sets; N6-(5-carboxypentyl)adenine selected aptamers, the Arabidopsis genome/transcriptome, and cytokinin microarray expression data are presented in A) as a Venn Diagram. Genes up- or down-regulated by cytokinin at least two-fold are summarized in light orange. Genes with a match to seven out of ten nucleotides in the N6-(5-carboxypentyl)adenine selected aptamer conserved core domain (underlined in B) are in light blue or green. Due to the short query sequences (each aptamer was 50 nt), and having few bases conserved outside the core, it was not possible to narrow search results using statistical cut-offs. Rather, low stringency search parameters were used (Expect Threshold: 1000, Word Size: 7, Match/Mismatch: 1,-3) to identify CaAps (Expect Threshold: 1000, Word Size: 7, Match/Mismatch: 1,-3). The first reason for this was to retrieve all possible binding sequences for further analyses. The second reason is because while the ligand-binding core is probably conserved, other nucleotides in the riboswitch need only form proper base-pairing structures and could have higher sequence heterogeneity. Both the non-redundant nt collection and RefSeq mRNA databases were searched limiting results to Arabidopsis thaliana (taxid: 3702). Approximately 100 nucleotides on either side of the CaAp sequences were retrieved from the NCBI (National Center of Biotechnology Information) core nucleotide entry. NCBI and TAIR (The Arabidopsis Information Resource) databases were used to map sequences to a location in the Arabidopsis transcriptome and classified as Coding (light blue) or non-coding (light green). One gene At4g23180, CRK10 (underlined in A) was selected as an illustration of the hypothesis because it is down-regulated three-fold by cytokinin, it is in an intron, and, as depicted in B) its sequence is conserved with the 5aA2 aptamer [28] throughout, and especially in the core domain (underlined).