Figure 7.
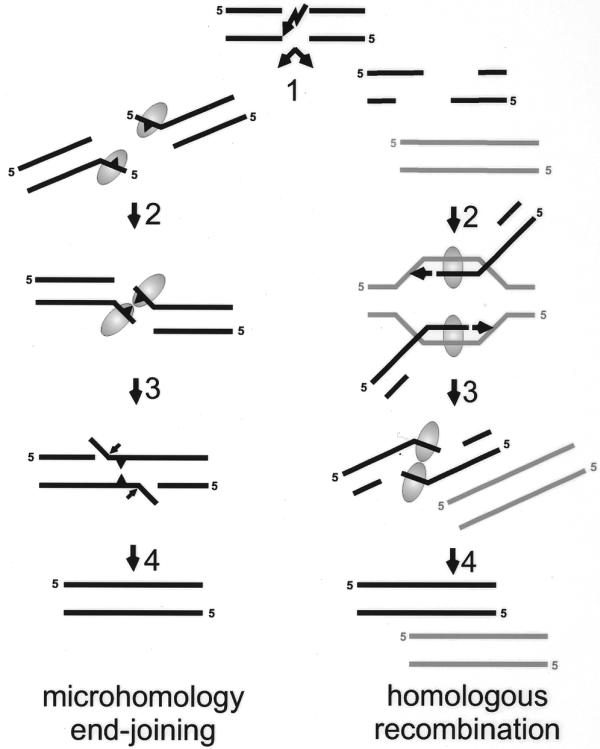
Proposed models for the possible functions of the Mre11 complex in end-joining and homologous recombination. If a DSB is repaired via microhomology-directed end-joining, both ends can be either nucleolytically processed or partially unwound (left, step 1: microhomologies symbolized by triangle). The Mre11-containing complex (symbolized by the oval) may bind the short ssDNA tails. The tails could be kept in close proximity through interaction between two Mre11 protomers either directly or as constituents of the same Rad50/Mre11 complex (left, step 2). The single-strand-annealing activity of Mre11 promotes base pairing between complementary DNA sequences (left, step 3). In the last step the activities of processing enzymes such as flap-endonucleases, polymerases and ligase(s) are required to restore the continuity of the DNA molecule (left, step 4). If the DSB is repaired via homologous recombination, extensive degradation of both ends results in long 3′ ssDNA tails (right, step 1). After strand invasion and homologous pairing (right, step 2) by the Rad52 epistasis group proteins and RPA (not indicated), new DNA is synthesized (arrowhead). Restoration of the lost sequence information will be complete when both newly-synthesized strands overlap (right, step 3). A possible resolution step of this recombination intermediate would involve unwinding of the newly-synthesized strand from the template DNA followed by annealing of the complementary newly-synthesized strands, possibly by Mre11. Finally, single-strand gaps are filled in and ligated, completing the repair of the damaged DNA molecule (right, step 4).
