Figure 3.
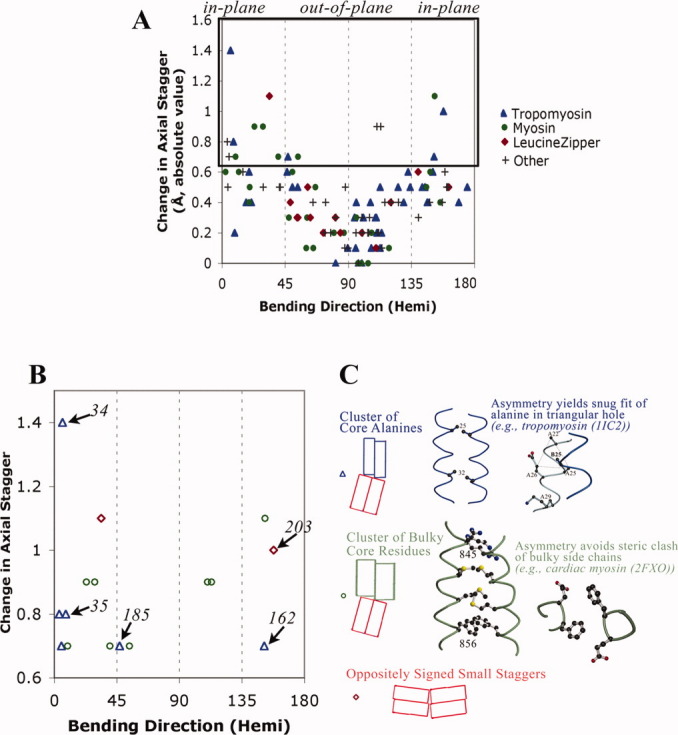
Large changes in axial staggering of the α-helices promote in-plane bending. A: Scatter plot of results from summary log files of all relatively high resolution crystal structures of parallel homodimeric coiled coil according to SCOP8 as of January 15, 2009. In addition to tropomyosin (see Fig. 2), coiled coils are from myosin (pdb i.d. 1nkn; 3bas; 3bat; 2fxm; 2fxo), leucine zipper domain (2zta, 1zik, 2ahp, 1zil, 1pyI, 1zme, 1hwt, 2hap, 1qp9, 1gd2, 1gu4, 1gtw, 1h8A, 1h88, 1hjb, 1io4, 1nwq, 2c9l, 2c9n), and other proteins (1dkg, 1d7m, 1deb, 1joc, 1no4, 1noh, 1uix, 1s1c, 1x79, 1tu3, 1T6F, 1uii, 2gzd, 2gzh, 2d7c, 2hv8, and 2ocy) (see also Table SIII). The hemicircle-reduced direction of bending is shown on the x-axis. At each position along the sequence, i, the change in axial stagger per heptad = [axial stagger at position (i+3) + stagger at position (i+4) − stagger at (i−3) − stagger at (i−4)]/2.0, and the absolute value is printed out on the y-axis. B: The 16 bends with greatest change in axial staggering. Italicized numbers are tropomyosin residues. C: Two major designs that promote axial staggering. See also text.
