Abstract
B cell maturation and B cell-mediated antibody response require programmed DNA modifications such as the V(D)J recombination, the immunoglobulin (Ig) class switch recombination, and the somatic hypermutation to generate functional Igs. Many protein factors involved in DNA damage repair have been shown to be critical for the maturation and activation of B cells. Rad9 plays an important role in both DNA repair and cell cycle checkpoint control. However, its role in Ig generation has not been reported. In this study, we generated a conditional knock-out mouse line in which Rad9 is deleted specifically in B cells and investigated the function of Rad9 in B cells. The Rad9−/− B cells isolated from the conditional knock-out mice displayed impaired growth response and enhanced DNA lesions. Impaired Ig production in response to immunization in Rad9−/− mice was also detected. In addition, the Ig class switch recombination is deficient in Rad9−/− B cells. Taken together, Rad9 plays dual roles in generating functional antibodies and in maintaining the integrity of the whole genome in B cells.
Keywords: Antibodies, Apoptosis, Cell Cycle, Cell Division, DNA Repair, B Cells, Rad9, Class Switch Recombination, Genome Integrity, Proliferation
Introduction
The immune cells, especially B cells, are exposed to various genotoxic stresses during their maturation and activation for immune responses. The DNA damages in B cells are mainly the results of programmed mechanisms, such as the V(D)J recombination, the class switch recombination (CSR),2 and the somatic hypermutation of the Ig gene (1). Both V(D)J recombination and CSR can specifically generate DNA double strand breaks (DSBs), and DSB is a particularly harmful form of DNA damage. Somatic hypermutation introduces point mutations, and occasionally deletion or insertion mutations, into the rearranged V(D)J gene segments at a frequency of ∼10−3 mutations per base per round of cell division. Besides these three specific DNA-altering mechanisms, B cells are also exposed to general DNA injuries as they rapidly proliferate in response to stimuli (1, 2). Although the aforementioned DNA DSBs and mutations are required for normal immune response, these DNA lesions, if not managed appropriately, can lead to severe consequences such as immune deficiency or cancer (3).
Although several of the common DNA damage response mechanisms are found to be responsible for maintaining the genome integrity of immune cells, some of the DNA damage response pathways play unique roles in B cells(3). For example, in germinal center B cells, some DNA repair factors involved in mismatch repair (MMR) or base excision repair (BER) are diverted from their normal roles in preserving genomic integrity to increasing DNA sequence diversity within the Ig locus (4).
Rad9 plays important roles in both cell cycle checkpoint control and DNA repair (5–7). Rad9 is evolutionarily conserved from yeast to humans and can form a ring-shaped heterotrimer, dubbed the 9-1-1 complex, with Rad1 and Hus1 (8–11). Its deletion in the fission yeast Schizosaccharomyces pombe inactivates S/M, intra-S, and G2/M checkpoint controls and sensitizes fission yeast cells to killing by UV light, γ-rays, and the replication inhibitor hydroxyurea (12–14). Disruption of the mouse ortholog of Rad9 also leads to impaired S/M and G2/M checkpoint controls and sensitizes mouse cells to UV light, γ-rays, and hydroxyurea (15). Aside from cell cycle checkpoint functions, there is mounting evidence that Rad9 has important roles in repairing DNA lesions. Rad9 can bind multiple DNA repair proteins involved in DNA BER and regulate their activities (16–23). Recently, we reported that Rad9 carries out its MMR function through interaction with MLH1 (24). Bai et al. (25) reported that Rad9 could also physically and functionally interact with the other two MMR proteins, MSH2 and MSH6. Interestingly, both BER and MMR are required specifically in Ig production (4, 26).
Here, to test the possible roles of Rad9 in B cells, we generated a conditional knock-out mouse line in which Rad9 is deleted specifically in B cells. Mice with Rad9+/− or Rad9−/− B cells demonstrate no overt, spontaneous, morphologic defects. However, the Rad9−/− B cells displayed impaired growth response and enhanced DNA lesions. We also detected impaired Ig production in response to TNP-KLH immunization in Rad9−/− mice. In addition, the Ig CSR is deficient in Rad9−/− B cells. These data demonstrate that Rad9 plays dual roles during B cell development in generating functional antibodies and in maintaining the integrity of the whole genome.
EXPERIMENTAL PROCEDURES
Generation of Rad9Tar/TarCD19cre/+ Mice
B cell-specific and Rad9-deficient mice were generated by crossing Rad9Tar/Tar 129SvEv strain mice (15) with CD19-Cre knock-in C.129P2-Cd19tm1(cre)Cgn/J strain mice expressing Cre under control of the endogenous CD19 promoter (The Jackson Laboratory, Bar Harbor, ME). Methods for PCR genotyping of mouse tissues as well as isolated cells for the Rad9-loxP loci and Cre-mediated recombination were identical to procedures previously described (15). To detect the presence of the targeted sequence, primers 5′-TTCGGGTGGGAGAATCAGAC-3′ (T1) and 5′-GGATCTCTCCCCATTCACCA-3′(T2) were used. To detect the presence of the first two exons of Rad9, primers 5′-CCGGGTGAACCAATAAGGAA-3′ (D1) and 5′-AAGGAAGCAGGCATAGGCAG-3′ (D2) were used (see Fig. 1A). The animal-handling protocols and the following procedures were approved by the Institutional Animal Care and Use Committee at the Institute of Biophysics, Chinese Academy of Sciences.
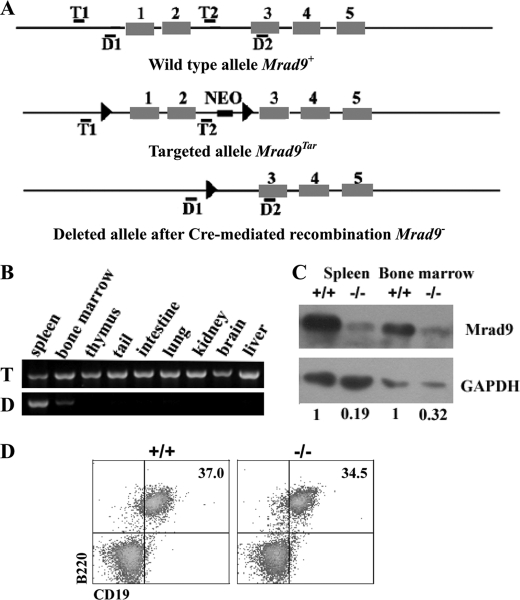
FIGURE 1.
Rad9 deletion in B cells. A, maps of original, targeted, and deleted Rad9 genomic DNA fragments. Black boxes represent exons, and thin lines represent introns as well as DNA sequences surrounding Rad9 gene. Locations of primer pairs for detecting the targeting (T1/T2) and deletion (D1/D2) of the first two exons are marked. B, PCR genotyping of Rad9 deletion in mouse tissues. The deleted signature DNA band was only observed in spleen and bone marrow. T and D on the left of PCR panels indicate targeted and deleted signature bands of Rad9, respectively. C, Western blotting analyses for Rad9 protein levels in B cells purified from spleen and bone marrow. Numbers reflect the intensity of bands representing Rad9 protein levels normalized to GAPDH in Rad9−/− and Rad9+/+ B cells. D, flow cytometric analysis of CD19 expression. Cells isolated from the spleen of 6–8-week-old mice were stained with the antibodies indicated in the figure. Numbers show relative percentages of cells within the indicated gates. These analyses are averages of three pairs of Rad9+/+ and Rad9−/− mice. +/+ and −/− represent Rad9+/+ and Rad9−/− genotypes, respectively.
B Cell Isolation
6-to-8-week-old mice were sacrificed. Cells isolated from bone marrow and spleen were treated with 0.83% NH4Cl/Tris-HCl (pH 7.2) to lyse red blood cells. B cells were collected by sorting B220-positive cells using a dual laser FACSVantageTM (BD Biosciences). The cells were>95% B220+, as verified by flow cytometry.
Western Blotting
Western blotting was carried out as described previously (27). Primary and secondary antibodies used in this study are mouse anti-RAD9 (BD Biosciences), mouse anti-GAPDH (KangChen, Shanghai, China), and peroxidase-conjugated anti-mouse IgG (Sigma).
Flow Cytometric Analysis
Lymphoid cells stained with anti-B220-PE, anti-CD19-FITC, anti-B220-FITC, anti-CD23-FITC, anti-CD21-PE, anti-IgD-PE, anti-CD43-PE, anti-IgM-APC (Allophycocyanin) (Pharmingen), and anti-IgG-CyTM5 (Jackson ImmunoResearch Laboratories, West Grove, PA) were analyzed by FACSCalibur cytometer (BD Biosciences).
In Vitro Expansion Assay
Purified splenic B cells (106 cells/ml) at 2 × 105 cells/well in triplicate in 96-well plates were cultured in RPMI medium 1640 containing 50 μm 2-mercapitoethanol, 2 mm l-glutamine, 100 μg/ml penicillin, 100 μg/ml streptomycin, 1 mm sodium pyruvate, 15 ng/ml IL-4 (PeproTech, Rocky Hill, NJ), and 25 μg/ml LPS (Sigma). Cell proliferation was evaluated by cell counting, and viability was analyzed by trypan blue exclusion assay every 24 h.
BrdU Uptake Assays
Purified splenic B cells (106 cells/ml) were cultured in medium with IL-4 and LPS for 2 days. 10 μm BrdU was added to medium, and cells were pulse-labeled for 40 min. Cells were then processed and probed with FITC-conjugated anti-BrdU antibody (BD Biosciences) and stained with propidium iodide (PI). Flow cytometric analyses were performed on a FACSCalibur.
Apoptosis Assays
The purified splenic B cells stimulated with IL-4 and LPS for 2 days were washed twice with cold PBS and then resuspended in 1× binding buffer (10 mmol/liter HEPES, 140 mmol/liter NaCl, and 2.5 mmol/liter CaCl2) at a concentration of 1 × 106 cells/ml. Then cells were stained with annexin V-FITC (Jingmei Biotech, Shenzhen, Guangdong, China) and PI for 15 min at room temperature and subjected to flow cytometric analysis.
Neutral Comet Assay
The neutral comet assay is used for detecting DNA double strand breaks. The purified splenic B cells were stimulated with IL-4 and LPS for 2 days. The comet assay was carried out according to the manufacturer's instructions (Trevigen, Gaithersburg, MD). Briefly, cells at a concentration of 1 × 105 cells/ml were mixed gently with premelted low temperature-melting agarose at a volume ratio of 1 to 10 (v/v) and spread on glass slides. The slides were then submerged in precooled neutral lysis buffer at 4 °C for 30 min. After rinsing, the slides were equilibrated in Tris borate-EDTA solution, electrophoresed at 1.0 V/cm for 20 min, and then stained with PI. Fluorescence images for at least 50 nuclei were captured using a Nikon microscope and analyzed by CASP version 1.2.2 software (University of Wroclaw) for tail moment (the geometric mean of fluorescence on the tail from the nucleus).
Immunization and ELISA
Mice were immunized intraperitoneally with T cell-dependent antigene TNP-KLH (20 μg/mouse; Biosearch Technologies, Novato, CA) in Freund's complete adjuvant. At the day 14 after immunization, the sera were collected and assayed by TNP-specific and isotype-specific ELISA to determine the levels of the antigen-specific immune response. Briefly, serial dilutions of the immune sera were incubated on 96-well microtiter plates coated with TNP-BSA (Biosearch Technologies) and probed with horseradish peroxidase-conjugated rabbit anti-mouse isotype-specific IgM, IgG, IgG1, and IgG3 (SouthernBiotech, Birmingham, AL) and the substrate O-phenylenediamine dihydrochloride, and the results were read at 492 nm.
Measurement of IgG Production in Vitro
Red blood cell-depleted total splenocytes were labeled with CFSE (5 μm, Invitrogen) and cultured for 4 days in RPMI medium 1640 and supplements as mentioned above. The cells were harvested and stained with Cy5-labeled monoclonal rat anti-mouse IgG and analyzed using a FACSCalibur cytometer. B cells were identified by anti-B220-PE antibody.
Statistical Analysis
The significance of the difference was calculated by Student's t test, and p < 0.05 was considered statistically significant.
RESULTS
Targeted Deletion of Rad9 in Mouse B Cells
To determine the functional involvement of Rad9 in B cells, we deleted the Rad9 gene specifically in B cells by conditional gene targeting using Cre/loxP-mediated recombination. The mice containing loxP targeted to Rad9 were crossed with mice in which Cre transcription is under the control of the CD19 promoter (28). CD19 is a B cell-specific gene and expressed through all B cell developmental stages, and thus Cre-mediated deletion of the Rad9 gene was expected to occur only in B cells. Indeed, deletion of Rad9 took place exclusively in tissues (spleen and bone marrow) that contain B cells (Fig. 1B). Additionally, Western blotting illustrated that as compared with Rad9+/+ B cells, Rad9 protein expression was reduced by 81 and 68% in Rad9−/− B cells purified from the spleen and the bone marrow, respectively (Fig. 1C). The Rad9 deletion in splenic B cells is obviously more efficient than that in bone marrow B cells. The above results indicate that the Rad9 was deleted specifically in B cells. We observed that Rad9 was not completely deleted in spleen and bone marrow B cells. This phenomenon was also reported by other groups using the same model system to delete other genes including those playing important roles in both DNA repair and Ig generation (28, 29).
CD19 is a hallmark differentiation antigen of the B lineage. It has been reported that mice hemizygous for the cre insertion (CD19cre/+), which retain one functional CD19 allele, are phenotypically normal and can be used for B lineage-specific deletion of a floxed target gene (28). In this study, we also detected CD19 expression on spleen B cells using flow cytometry. The expression of CD19 on B cells was not affected in Rad9−/− mice as compared with Rad9+/+ mice in the spleen (Fig. 1D). Therefore, in this model, there is no difference in CD19 expression in Rad9−/− mice and Rad9+/+ mice, and this excludes the possibility that different CD19 expression in Rad9−/− mice and Rad9+/+ mice contributes to the unique phenotype in Rad9−/− B cells. For the purpose of concise description, Rad9+/+, Rad9+/− and Rad9−/− will be used to denote Rad9+/+CD19cre/+, Rad9Tar/+CD19cre/+, and Rad9Tar/Tar CD19cre/+, respectively, in the rest of this study.
Effect of Rad9 Knock-out on B Cell Maturation
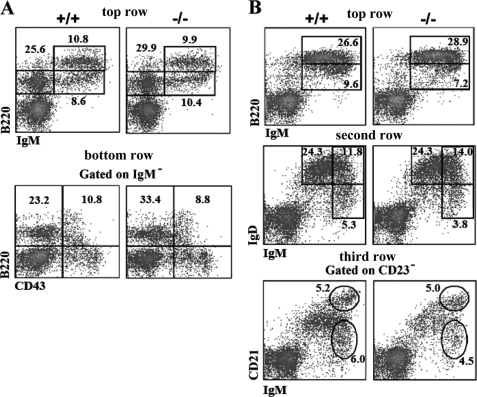
Rad9-null mutation causes mouse embryonic lethality, whereas no obvious defects were observed in mice with a specific deletion of Rad9 in keratinocytes (15, 27). Similarly, in this study, mice with Rad9+/− or Rad9−/− B cells showed no overt defects in gross morphology or health up to 1.5 years of age (12 Rad9−/−, 8 Rad9+/−, and 11 wild-type mice were maintained and carefully observed for any abnormal signs for 1.5 years). We analyzed the maturation of B cells in Rad9+/+ and Rad9−/− mice using flow cytometry with various B lineage differentiation markers, and our results showed that both Rad9+/+ and Rad9−/− B cells from mouse bone marrow and spleen developed normally. There were no significant differences in the percentage of different subsets of B cells, including pro-B (IgM−B220+CD43+), pre-B (IgM−B220+CD43−), immature B (IgM+B220lo), and mature B (IgM+B220hi) of the bone marrow and follicular B cells (IgMloIgDhi), transitional type-2 B cells (IgMhiIgDhi), marginal zone B cells (IgMhiIgDlo and CD23−IgMhiCD21hi), and transitional type-1 B cells (IgMhiIgDlo and CD23−IgMhiCD21lo) of the spleen (Fig. 2). Therefore, although Rad9 is essential for embryogenesis, its deletion has no significant effect on general B cell development.
FIGURE 2.
Flow cytometric analysis of lymphoid cell development of Rad9+/+ and Rad9−/− mice. Cells isolated from the bone marrow (A) and spleen (B) of 6–8-week-old mice were stained with the antibodies indicated in each figure. Numbers show relative percentages of cells within indicated gates. These analyses are averages of three pairs of Rad9+/+ and Rad9−/− mice. A, in the bone marrow (BM), top row, staining for pro/pre-B (IgM−B220+), immature (IgM+B220lo), and mature recirculating (IgM+B220hi) B cells; bottom row, staining for pro-B cells (IgM−B220+CD43+) and pre-B cells (IgM−B220+CD43−). B, in the spleen, top row, staining for immature (IgM+B220lo) and mature (IgM+B220hi) B cells; second row, staining for follicular B cells (IgMloIgDhi), transitional type-2 B cells (IgMhiIgDhi), and marginal zone/transitional type-1 B cells (IgMhiIgDlo); third row, staining for transitional type-1 B cells (CD23−IgMhiCD21lo) and marginal zone B cells (CD23−IgMhiCD21hi).
Rad9-dependent B Cell Expansion, Apoptosis, and Genomic Integrity in Response to Mitogenic Stimuli
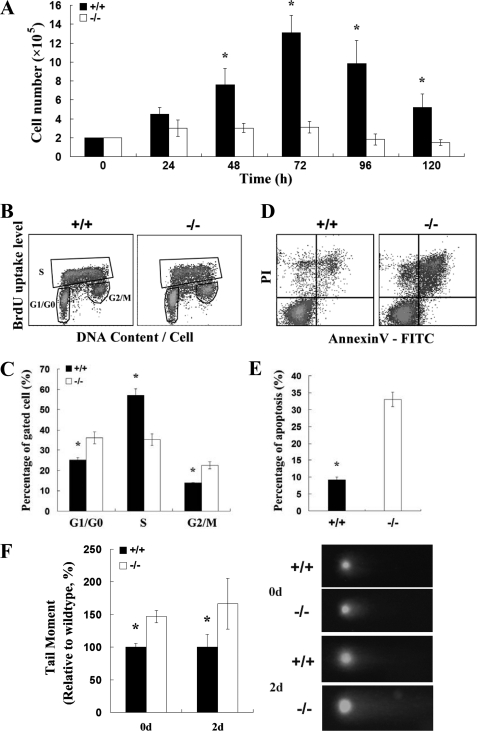
B cells undergo clonal expansion in response to stimuli. To evaluate the response of Rad9+/+ and Rad9−/− B cells, purified splenic B cells were cultured with IL-4 (15 ng/ml) and LPS (25 μg/ml). Cell proliferation was analyzed by counting viable cells every 24 h in culture. The number of wild-type cells increased significantly with the peak at 72 h after stimulation (6.6 times the seeded level). In contrast, the number of Rad9−/− B cells only slightly increased after stimulation with the peak at 72 h (1.6 times the seeded level, Fig. 3A). The impaired Rad9−/− B cell expansion may be caused by impaired cell proliferation or increased cell death. We further analyzed cell proliferation by examining DNA replication and DNA content using BrdU and PI after 2 days of stimulation. As compared with wild-type B cells, there was significant reduction in the number of BrdU-positive S phase Rad9−/− B cells (35.34 versus 56.92%, p < 0.01, n = 3, Fig. 3, B and C). The measurement of BrdU uptake in combination with PI staining in individual cells provides the information on cell cycle distribution. Using this method, we found a higher number of Rad9−/− B cells at G1/G0 phase than of Rad9+/+ B cells (36.12 versus 25.24%, p < 0.05, n = 3, Fig. 3, B and C). Furthermore, there was a greater number of Rad9−/− B cells at G2/M phase than of Rad9+/+ B cells (22.46 versus 13.9%, p < 0.05, n = 3, Fig. 3, B and C). These results suggest that Rad9 deficiency causes cell cycle slowing down or rest in G1/G0 and G2/M phases, which lead to a hypoproliferative response to the stimulation of IL-4 and LPS. To determine the effect of Rad9 deficiency on B cell apoptosis, we stained cells with annexin V-FITC and PI. We found that the percentage of Rad9−/− B cells that underwent apoptosis was significantly higher than that of Rad9+/+ B cells after 2 days of stimulation (32.19 versus 8.57%, p < 0.01, n = 3, Fig. 3, D and E). Thus, the reduction in the number of Rad9−/− B cells was a result of both impaired cell proliferation and increased apoptosis.
FIGURE 3.
Rad9-dependent B cell expansion, apoptosis, and genomic integrity in response to LPS plus IL-4 in vitro. A, kinetics of the responses of Rad9+/+ and Rad9−/− B cells to LPS and IL-4 are similar with the peak at 72 h after stimulation. Cell proliferation was analyzed by counting trypan blue-excluded viable cells at the indicated times. The results are averages of three experiments. B, flow cytometric analysis of B cells stained with both PI and BrdU after 2 days of incubation. BrdU-positive S phase cells, BrdU-negative G1/G0 phase cells, and BrdU-negative G2/M phase cells were gated. C, quantitative comparison of cell cycle distribution between Rad9+/+ and Rad9−/− B cells. Three measurements as shown in B were carried out for comparison. D, flow cytometric analysis of B cells to assess spontaneous apoptosis using annexin V labeling. The B cells stimulated with IL-4 and LPS were incubated for 2 days before harvesting for apoptotic analysis. E, quantitative comparison of apoptosis between Rad9+/+ and Rad9−/− B cells. Three apoptotic assays shown in D were carried out for comparison. F, evaluation of DNA double strand breaks in resting B cells (day 0) and in B cells after 2 days of incubation by the neutral comet assay. On the right, representative comet assay results were shown, and on the left, quantitative comparison of comet tail moments between 100 Rad9−/− B cells and 100 Rad9+/+ B cells are shown. Student's t test, *, p < 0.05. Error bars represent the mean ± S.E.
Because Rad9 plays an important role in DNA damage repair (5–7), the accumulations of Rad9−/− B cells in G1/G0 and G2/M phase and a higher number of apoptotic Rad9−/− B cells might be caused by an increase in DNA lesions. To confirm the effects of Rad9 deficiency on DNA damage response, we quantified DNA DSBs in Rad9+/+ and Rad9−/− B cells before and after 2 days of stimulation using the neutral comet assay. There were significantly more DSBs in Rad9−/− B cells than in Rad9+/+ B cells before and after stimulation (Fig. 3F). Stimulation of IL-4 and LPS enhanced DSBs in both Rad9+/+ and Rad9−/− B cells as expected. Therefore, Rad9 is critical for maintaining genomic integrity in B cells. The higher level of DSBs likely contributes to heightened apoptosis of Rad9−/− B cells. Taken together, the results suggest that maintaining genomic integrity of B cells depends on Rad9 and is important for clonal expansion response of B cells (27).
Defective CSR and Impaired Humoral Immune Response in Rad9−/− Mice
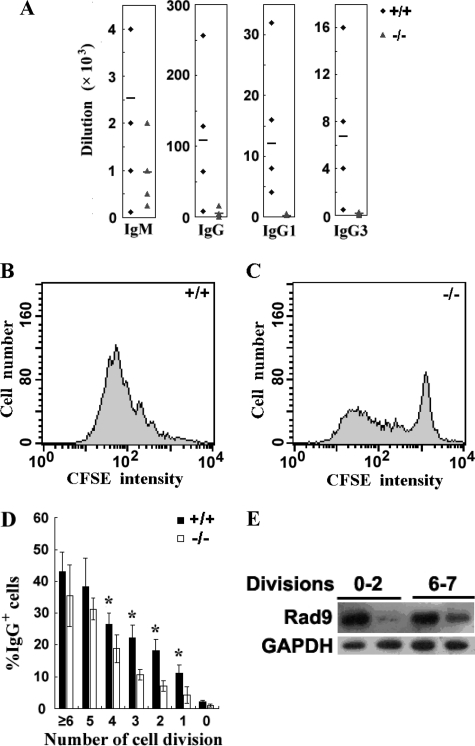
To determine the role of Rad9 in humoral response, we compared the level of antigen-specific IgM, IgG, and IgG subclasses in the blood of Rad9−/− and Rad9+/+ mice after intraperitoneal immunization with TNP-KLH in the Freund's complete adjuvant using ELISA. The average titers of TNP-specific IgM in Rad9+/+ mice and Rad9−/− mice were 1:2520 and 1:958, respectively (p = 0.08, n = 6), a 2.63-fold decrease in Rad9−/− mice. The average titers of TNP-specific IgG produced by Rad9+/+ mice and Rad9−/− mice were 1:108,000 and 1:4208, respectively (p < 0.05, n = 6), a 25.7-fold decrease in Rad9−/− mice. Furthermore, the average titers of both IgG1 and IgG3 were significantly decreased in Rad9−/− mice as compared with the Rad9+/+ mice (Fig. 4A). These results demonstrate a defective humoral response against a T cell-dependent antigen in Rad9−/− B cells.
FIGURE 4.
Impaired humoral immune response of Rad9−/− mice and defective CSR in Rad9−/− B cells. A, serum TNP-specific Ig levels in Rad9+/+ and Rad9−/− mice at 14 days after immunization with TNP-KLH were determined by ELISA. Data are plotted as the dilution in six mice of each genotype. Diamonds represent the values derived from Rad9+/+, and triangles represent the values derived from Rad9−/− mice. The bar denotes the mean of each group. B and C, cell divisions shown by CFSE dye dilution of Rad9+/+ and Rad9−/− B cells stimulated with LPS and IL-4 for 4 days were analyzed by flow cytometry. D, flow cytometric analysis of surface IgG expression by CFSE-labeled Rad9+/+ and Rad9−/− B cells after 4 days of LPS and IL-4 stimulation. The percentage of cells expressing IgG is calculated from and shown separately for each population that has undergone the indicated number of cell divisions. The data represent the mean ± S.E. of four cell cultures. E, Western blotting analyses for Rad9 protein levels in B cells with different cell divisions. We collected B cells that had completed 0–2 cell divisions and six cell divisions as well as more than 6 cell divisions by cell sorting and measured Rad9 protein levels by Western blotting. The cells that survived six cell divisions or more than six cell divisions expressed higher levels of Rad9 protein than the cells that survived 0–2 cell divisions. Student's t test, *, p < 0.05.
The profound reduction of IgG production in Rad9−/− mice raises a possibility that Rad9 participates in CSR. CSR requires cell proliferation, and we have shown that Rad9 is required for cell proliferation (Fig. 3A). Because the effect of Rad9 deletion on B cell proliferation is much smaller (6.6/1.6 = 4.13-fold decrease) than on IgG generation (25.7-fold decrease), we inferred that Rad9 played a more direct role in CSR. Here, we determined whether there was a significant reduction of IgG in Rad9−/− cells that pass through the same cell proliferation stages. CFSE labeling can indicate the number of cell divisions through which a cell is derived from an original cell because the CFSE is decreased proportionally with the number of cell divisions. We used CFSE labeling and flow cytometry to determine the number of cell cycle divisions and the appearance of IgG-expressing B cells simultaneously as cells proliferated. After 4 days in culture with IL-4 and LPS, the percentage of B cells expressing IgG in the cells underwent different numbers of cell divisions according to CFSE staining was determined (Fig. 4, B–D). We found that in cells at the same cell cycle divisions (the same levels of intracellular CFSE staining), IgG expression levels in B cells from Rad9−/− mice were significantly lower than those in Rad9+/+ B cells (Fig. 4D). The average IgG-positive B cells at different numbers of cell divisions of Rad9−/− and Rad9+/+ mice were 4.35 versus 11.14% (p < 0.01, n = 4) at D1 (after the first division), 7.04 versus 18.30% (p < 0.01, n = 4) at D2, 10.62 versus 22.27% (p < 0.01, n = 4) at D3, 18.75 versus 26.44% (p < 0.05, n = 4) of D4, 31.26 versus 38.14% (p > 0.05, n = 4) at D5, and 35.54 versus 43.00% (p > 0.05, n = 4) at D6 and cell divisions after D6. These results indicate that during the first four division periods, the IgG expression in individual Rad9−/− B cells is statistically significantly lower than that in individual Rad9+/+ B cells, suggesting that Rad9 plays a more direct role in CSR than just through cell expansion.
The decrease in cell surface IgG was more apparent in cells that had undergone fewer cell divisions (Fig. 4D). The possible explanation is that cells with residual Rad9 protein may have a selective advantage in both survival and CSR. To test this hypothesis, we purified B cells that had completed 0–2 cell divisions and cells that had completed six divisions as well as more than six cell divisions by cell sorting, and Rad9 protein levels of the cell lysates were measured by Western blotting. We found that cells that survived six or more than six cell divisions expressed much higher levels of Rad9 protein than those that survived 0–2 cell divisions, although Rad9 protein levels in Rad9−/− cells were lower than in Rad9+/+ cells in both cell division ranges (Fig. 4E). This result confirms that Rad9 plays a critical role in CSR.
DISCUSSION
In this study, we have documented that Rad9 is required for B cell proliferation (Fig. 3), Ig generation, and CSR (Fig. 4). Accumulated evidence indicates that Rad9 plays an important role in DNA repair (5–7), and enhanced DNA lesions resulting from Rad9 deletion in mouse keratinocytes lead to enhanced apoptosis and a slower cell cycle progression (27). Consistently, we also observed enhanced levels of DNA lesions (Fig. 3F) and apoptosis (Fig. 3, D and E) and a slower cell cycle progression (Fig. 3, B and C) in Rad9-deleted mouse B cells. The total effect of the enhanced apoptosis and a slower cell cycle progression is a reduced mouse Rad9−/− B cell proliferation (Fig. 3A). Therefore, the reduced proliferation rate is one of the mechanisms underlying deficient Ig generation in Rad9−/− B cells (Fig. 4A).
In Rad9−/− mice, the reduction of blood TNP-specific IgG was much more significant than that of blood TNP-specific IgM (25.7-fold decrease versus 2.63-fold decrease: Fig. 4A), suggesting that a CSR deficiency resulted from Rad9 deletion. In addition, in cells at the same cell cycle divisions, IgG expression levels in B cells from Rad9−/− mice were significantly lower than those in Rad9+/+ B cells (Fig. 4, B–D), suggesting that Rad9 plays a more direct role in Ig production and CSR than just through cell expansion.
CSR is an intrachromosomal deletional recombination event that occurs in mature B cells in response to antigen stimulation. It has been reported that many DNA damage repair factors required for BER, MMR, and nonhomologous end joining directly participate in this process (4, 26). Accumulated evidence indicates that Rad9 plays direct roles in base excision repair and mismatch repair, but not in nonhomologous end joining of DNA double strand breaks (23–25, 30). We previously reported that Rad9 directly interacted with the MMR protein hMLH1, and this interaction is important for DNA MMR (24). Bai et al. (25) have reported recently that Rad9 also physically and functionally interacts with the two other important MMR proteins, hMSH2-hMSH6. Rad9 can stimulate the DNA binding activity of hMSH2-hMSH6, and Rad9 and hMSH6 colocalize to nuclear foci in nuclei of HeLa cells after exposure to N-methyl-N′-nitro-N-nitrosoguanidine, which causes DNA lesions that can be repaired by the MMR mechanism. Furthermore, the foci formation of Rad9 and hMSH6 in N-methyl-N′-nitro-N-nitrosoguanidine-treated cells is dependent on the presence of each other. Gembka et al. (23) reported that Rad9 physically interacted with the BER protein APE1 and that the 9-1-1 (Rad9-Hus1-Rad1) complex stimulated the endonuclease activity of APE1. Interestingly, it has been reported that the MMR proteins MLH1, MSH2, and MSH6 and the BER protein APE1 all function in CSR. CSR is reduced in mice that lack one of these four genes (31–34). Therefore, Rad9 is likely to participate in the process of CSR through BER and/or MMR, but not through nonhomologous end joining.
Based on the accumulated evidence, we hypothesize two molecular mechanisms by which Rad9 participates in the process of CSR. First, Rad9 is likely to contribute to the recruitment of the components of CSR machinery mentioned above and stimulate their functions in CSR. Second, Rad9 has been reported to possess 3′-to-5′-nuclease activity without a known function (35), and 3′-to-5′-nuclease activity has been shown to be important for MMR (36). Therefore, Rad9 is likely to function in mismatch-dependent 3′-to-5′-excision, thus promoting the conversion of the single strand break to DSB in CSR, having a similar role as ExoI, which is an MMR-associated 5′-directed exonuclease in eukaryotic cells and plays a role in CSR (37). To understand the exact underlying molecular mechanism(s) by which Rad9 functions in CSR (such as via its interaction with MLH1, MSH2, MSH6, or APE1), further major effort is required.
In summary, we have provided experimental evidences for the first time that Rad9 plays dual roles during B cell development in generating functional antibodies and in maintaining the integrity of the whole genome in B cells.
This work was supported by National Natural Science Foundation of China Grant 30700407, National Natural Science Foundation of China Grant 30530180, and National Protein Project of Ministry of Science and Technology Grant 2006CB910902.
- CSR
- class switch recombination
- BER
- base excision repair
- DSB
- double strand break
- KLH
- keyhole limpet hemocyanin
- TNP
- 2,4,6-trinitrophenol
- MMR
- mismatch repair
- PI
- propidium iodide
- CFSE
- carboxyfluorescein succinimidyl ester
- PE
- phosphatidylethanolamine
- h
- human.
REFERENCES
- 1.Revy P., Buck D., le Deist F., de Villartay J. P. (2005) Adv. Immunol. 87, 237–295 [DOI] [PubMed] [Google Scholar]
- 2.Branzei D., Foiani M. (2005) Curr. Opin. Cell. Biol. 17, 568–575 [DOI] [PubMed] [Google Scholar]
- 3.Xu Y. (2006) Nat. Rev. Immunol. 6, 261–270 [DOI] [PubMed] [Google Scholar]
- 4.Peled J. U., Kuang F. L., Iglesias-Ussel M. D., Roa S., Kalis S. L., Goodman M. F., Scharff M. D. (2008) Annu. Rev. Immunol. 26, 481–511 [DOI] [PubMed] [Google Scholar]
- 5.Sancar A., Lindsey-Boltz L. A., Unsal-Kaçmaz K., Linn S. (2004) Annu. Rev. Biochem. 73, 39–85 [DOI] [PubMed] [Google Scholar]
- 6.Hartwell L. H., Weinert T. A. (1989) Science 246, 629–634 [DOI] [PubMed] [Google Scholar]
- 7.Paulovich A. G., Hartwell L. H. (1995) Cell 82, 841–847 [DOI] [PubMed] [Google Scholar]
- 8.Griffith J. D., Lindsey-Boltz L. A., Sancar A. (2002) J. Biol. Chem. 277, 15233–15236 [DOI] [PubMed] [Google Scholar]
- 9.Xu M., Bai L., Gong Y., Xie W., Hang H., Jiang T. (2009) J. Biol. Chem. 284, 20457–20461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sohn S. Y., Cho Y. (2009) J. Mol. Biol. 390, 490–502 [DOI] [PubMed] [Google Scholar]
- 11.Doré A. S., Kilkenny M. L., Rzechorzek N. J., Pearl L. H. (2009) Mol. Cell 34, 735–745 [DOI] [PubMed] [Google Scholar]
- 12.Murray J. M., Carr A. M., Lehmann A. R., Watts F. Z. (1991) Nucleic Acids Res. 19, 3525–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lieberman H. B., Hopkins K. M., Laverty M., Chu H. M. (1992) Mol. Gen. Genet. 232, 367–376 [DOI] [PubMed] [Google Scholar]
- 14.Enoch T., Carr A. M., Nurse P. (1992) Genes Dev. 6, 2035–2046 [DOI] [PubMed] [Google Scholar]
- 15.Hopkins K. M., Auerbach W., Wang X. Y., Hande M. P., Hang H., Wolgemuth D. J., Joyner A. L., Lieberman H. B. (2004) Mol. Cell. Biol. 24, 7235–7248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu A. L., Bai H., Shi G., Chang D. Y. (2006) Front. Biosci. 11, 3062–3080 [DOI] [PubMed] [Google Scholar]
- 17.Chang D. Y., Lu A. L. (2005) J. Biol. Chem. 280, 408–417 [DOI] [PubMed] [Google Scholar]
- 18.Toueille M., El-Andaloussi N., Frouin I., Freire R., Funk D., Shevelev I., Friedrich-Heineken E., Villani G., Hottiger M. O., Hübscher U. (2004) Nucleic Acids Res. 32, 3316–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedrich-Heineken E., Toueille M., Tännler B., Bürki C., Ferrari E., Hottiger M. O., Hübscher U. (2005) J. Mol. Biol. 353, 980–989 [DOI] [PubMed] [Google Scholar]
- 20.Wang W., Brandt P., Rossi M. L., Lindsey-Boltz L., Podust V., Fanning E., Sancar A., Bambara R. A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 16762–16767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smirnova E., Toueille M., Markkanen E., Hübscher U. (2005) Biochem. J. 389, 13–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu X., Shell S. M., Zou Y. (2005) Oncogene 24, 4728–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gembka A., Toueille M., Smirnova E., Poltz R., Ferrari E., Villani G., Hübscher U. (2007) Nucleic Acids Res. 35, 2596–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He W., Zhao Y., Zhang C., An L., Hu Z., Liu Y., Han L., Bi L., Xie Z., Xue P., Yang F., Hang H. (2008) Nucleic Acids Res. 36, 6406–6417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bai H., Madabushi A., Guan X., Lu A. L. (2010) DNA Repair (Amst.) 9, 478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stavnezer J., Guikema J. E., Schrader C. E. (2008) Annu. Rev. Immunol. 26, 261–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu Z.., Liu Y., Zhang C., Zhao Y., He W., Han L., Yang L., Hopkins K. M., Yang X., Lieberman H. B., Hang H. (2008) Cancer Res. 68, 5552–5561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rickert R. C., Roes J., Rajewsky K. (1997) Nucleic Acids Res. 25, 1317–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reina-San-Martin B., Nussenzweig M. C., Nussenzweig A., Difilippantonio S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 1590–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pandita R. K., Sharma G. G., Laszlo A., Hopkins K. M., Davey S., Chakhparonian M., Gupta A., Wellinger R. J., Zhang J., Powell S. N., Roti Roti J. L., Lieberman H. B., Pandita T. K. (2006) Mol. Cell. Biol. 26, 1850–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schrader C. E., Edelmann W., Kucherlapati R., Stavnezer J. (1999) J. Exp. Med. 190, 323–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schrader C. E., Guikema J. E., Wu X., Stavnezer J. (2009) Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 645–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z., Scherer S. J., Ronai D., Iglesias-Ussel M. D., Peled J. U., Bardwell P. D., Zhuang M., Lee K., Martin A., Edelmann W., Scharff M. D. (2004) J. Exp. Med. 200, 47–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guikema J. E., Linehan E. K., Tsuchimoto D., Nakabeppu Y., Strauss P. R., Stavnezer J., Schrader C. E. (2007) J. Exp. Med. 204, 3017–3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bessho T., Sancar A. (2000) J. Biol. Chem. 275, 7451–7454 [DOI] [PubMed] [Google Scholar]
- 36.Iyer R. R., Pluciennik A., Burdett V., Modrich P. L. (2006) Chem. Rev. 106, 302–323 [DOI] [PubMed] [Google Scholar]
- 37.Bardwell P. D., Woo C. J., Wei K., Li Z., Martin A., Sack S. Z., Parris T., Edelmann W., Scharff M. D. (2004) Nat. Immunol. 5, 224–229 [DOI] [PubMed] [Google Scholar]