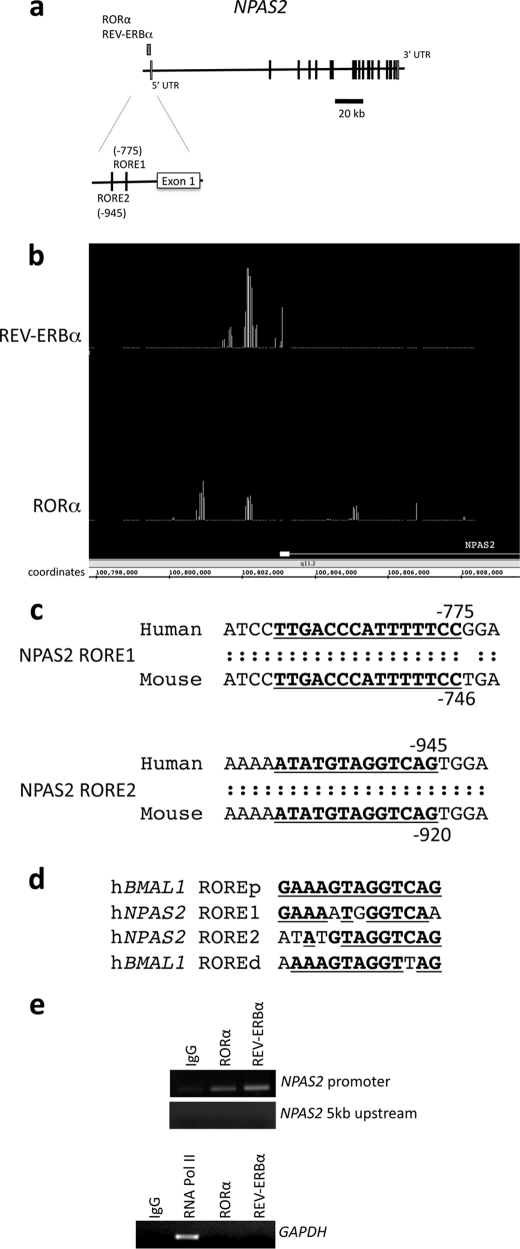
FIGURE 1.
Identification of REV-ERBα and RORα binding in the NPAS2 promoter using ChIP/chip assay. Both proteins bound to a region within 1 kb of the NPAS2 gene. a, schematic detailing the position of RORα and REV-ERBα occupancy within the promoter of the NPAS2 gene, as determined by ChIP/chip. The structure of the NPAS2 gene is shown, and exons and introns are indicated. The position of the nuclear receptor occupancy is indicated above the gene structure. b, screen image of primary ChIP/chip data from Integrated Genome Browser comparing the position of positive occupancy signals from the REV-ERBα and RORα experiments. The first exon of NPAS2 is illustrated in the figure, and the coordinates within chromosome 2 are also indicated. c, analysis of RORα and REV-ERBα binding region using the Evolutionarily Conserved Region browser, which showed two conserved response elements in the region. The sequence of the two putative response elements is shown, with the conserved regions underlined. The RORE1 (proximal) and RORE2 (distal) sites are shown. d, sequences of the ROREs from the NPAS2 promoter are compared with the sequences of the ROREs (proximal and distal) from the BMAL1 promoter. The underlined boldface letters represent regions of identity with the proximal RORE derived from the BMAL1 gene. e, ChIP assay was used to verify that REV-ERBα and RORα occupancy of the NPAS2 promoter containing the ROREs. The regions encompassing the two ROREs within the NPAS2 promoter are illustrated in the top gel as well as a control region ∼5 kb upstream of this region. Additionally, a control using GAPDH primers is shown with an RNA polymerase II positive control. Numbering of the sites is relative to the transcriptional start site.