Abstract
The stereospecific oxidative degradation of uric acid to (S)-allantoin was recently shown to proceed via three enzymatic steps. The final conversion is a decarboxylation of the unstable intermediate 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline (OHCU) and is catalyzed by OHCU decarboxylase. Here we present the structures of Klebsiella pneumoniae OHCU decarboxylase in unliganded form and with bound allantoin. These structures provide evidence that ligand binding organizes the active site residues for catalysis. Modeling of the substrate and intermediates provides additional support for this hypothesis. In addition we characterize the steady state kinetics of this enzyme and report the first OHCU decarboxylase inhibitor, allopurinol, a structural isomer of hypoxanthine. This molecule is a competitive inhibitor of K. pneumoniae OHCU decarboxylase with a Ki of 30 ± 2 μm. Circular dichroism measurements confirm structural observations that this inhibitor disrupts the necessary organization of the active site. Our structural and biochemical studies also provide further insights into the mechanism of catalysis of OHCU decarboxylation.
Keywords: Decarboxylase, Enzyme Catalysis, Purine, Uric Acid, X-ray Crystallography, Allantoin, Klebsiella pneumoniae, OHCU, Ureide
Introduction
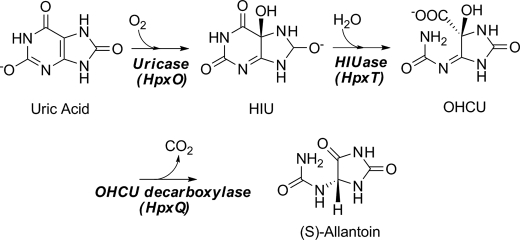
The ability to metabolize uric acid, a key intermediate in the degradation of purines, varies by organism. Humans, birds, reptiles, and some bacteria, for example, lack the enzymes necessary to break down this compound (1, 2). This inability to process uric acid in humans leads to high serum concentrations of urate that can crystallize in joints and cause gout. In plants and some bacteria, however, the high nitrogen content of the ureides make them attractive sources of nitrogen, particularly when other sources are limited. Organisms that possess the ability to degrade uric acid utilize a conserved pathway that converts this molecule into (S)-allantoin (1). Allantoin can be further degraded by plants and some bacteria for use as a nitrogen, carbon, and energy source.
The currently accepted degradative pathway from uric acid to allantoin is shown in Scheme 1. Until recently it was believed that only a single enzyme, urate oxidase, was responsible for the conversion of urate to allantoin. Over the last few years, however, several reports have conclusively proven that the pathway proceeds through two intermediates and utilizes two additional enzymes (3–5). The actual product of the urate oxidase reaction is the unstable intermediate, 5-hydroxyisourate (HIU).3 The conversion of HIU to another unstable compound, 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline (OHCU), is catalyzed by HIU hydrolase. OHCU can then be stereospecifically decarboxylated to yield (S)-allantoin.
SCHEME 1.
The catabolic pathway from uric acid to allantoin.
Despite recent efforts, the enzyme responsible for the final step in this pathway, OHCU decarboxylase, is not well characterized. Very little is known about the mechanism of this decarboxylation reaction, and only two structures of this enzyme, both from eukaryotes, have been solved and deposited in the Protein Data Bank. These two structures report a similar architecture for OHCU decarboxylase but display some distinct differences. Although both the structure from zebrafish (PDB code 2O70) (6) and the structure from Arabidopsis thaliana (PDB code 2Q37) (7) report dimeric structures, they differ in how the two protomers interact (6, 7). In addition, the structures reported with the bound product, allantoin, differ in which enantiomer is bound in the active site.
Beyond what has been inferred from the crystal structures, very few details about the kinetics or mechanism of the OHCU decarboxylase reaction are known. Sequence analysis of this enzyme reveals that it is unrelated to previously characterized decarboxylases, and biochemical analyses of the decarboxylation reaction by circular dichroism (CD) have shown that the reaction proceeds in the absence of exogenous cofactors. With the exception of a few relatively rare radical mechanisms, decarboxylation reactions generally require the presence of an electron sink or a leaving group to accommodate the electrons from the cleavage of the bond to the carboxylate moiety (8). This role is usually played by cofactors such as pyridoxal-5′-phosphate or thiamine pyrophosphate (8, 9). Few enzymes are known that catalyze decarboxylation reactions independent of cofactors. One notable example is orotidine 5′-monophosphate decarboxylase (OMPDC) (10, 11). This protein has been extensively studied in large part due to its extremely high catalytic proficiency (rate acceleration on the order of 1016) (12).
In this work we report the high resolution crystal structures of the cofactor-independent OHCU decarboxylase from Klebsiella pneumoniae, both in unliganded form and with bound allantoin. These structures represent the first example of a prokaryotic OHCU decarboxylase. Our structures reveal a prochiral, enol form of allantoin bound in the active site. This isomer more closely resembles the putative reaction intermediate than either of the individual isomers. The crystal structure with allantoin bound reveals an extensive hydrogen bonding network in the active site. Modeling of the substrate, putative reaction intermediate, and products provides additional details about the likely mode of catalysis. Utilizing the difference in absorbance of OHCU and allantoin at 254 nm, we characterize the steady state kinetic parameters for this enzyme. In addition, we have identified a novel competitive inhibitor of this enzyme and examined the method of inhibition using both x-ray crystal structures and circular dichroism measurements. The structure, modeling, and kinetic analysis allow us to propose a mechanism for the OHCU decarboxylase-catalyzed reaction.
EXPERIMENTAL PROCEDURES
Cloning and Mutagenesis
KpOHCU decarboxylase was expressed from a pET-28 based plasmid containing an N-terminal His6 tag and a tobacco etch virus cleavage site. The hpxQ gene was cloned from genomic DNA from K. pneumoniae subsp. pneumoniae (Schroeter) Trevisan MGH78578 (ATCC 700721). This construct, denoted HpxQ-THT, was provided by the Cornell Protein Production Facility and was cloned using standard molecular biology techniques. The Q88E mutant plasmid was made at the Cornell Protein Production Facility using site-directed mutagenesis of the native gene. Briefly, site-directed mutagenesis was performed on KpOHCU decarboxylase by a standard PCR protocol using Pfu Turbo DNA polymerase (Invitrogen) per the manufacturer's instructions and DpnI (New England Biolabs) to digest the methylated parental DNA before transformation. Clones were screened for the mutant by PCR with a screening primer and the T7 promoter primer. All clones and the mutant were verified by sequencing.
Protein Expression and Purification
BL21(DE3) cells were transformed with the HpxQ-THT plasmid and grown in LB media supplemented with 50 μg/ml kanamycin. A 10-ml starter culture, grown overnight, was used to inoculate 1 liter of LB cultures. These were grown at 37 °C with shaking until the A600 reached 0.4. The temperature was reduced to 20 °C, and after 30 min at this temperature, expression was induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside. The cells were grown for a further 16 h and then harvested by centrifugation and stored at −20 °C.
The cells were suspended in 50 mm Tris buffer, pH 7.6, containing 300 mm NaCl and 10 mm imidazole. After lysis by sonication, the cellular debris was removed by centrifugation, and the cleared lysate applied to a column containing pre-equilibrated nickel-nitrilotriacetic acid resin. The column was washed with 100 ml of 50 mm Tris buffer, pH 7.6, containing 300 mm NaCl, 25 mm imidazole, and 10% v/v glycerol. The protein was eluted with 50 mm Tris buffer, pH 7.6, containing 300 mm NaCl and 250 mm imidazole. The eluted protein was further purified by gel filtration on an ACTA Explorer FPLC with a HiLoad 26/60 Superdex prep grade G200 column running 10 mm Tris buffer, pH 7.6, with 30 mm NaCl. After purification, the protein was concentrated to 12 mg/ml using a centrifugal concentrator, and aliquots were flash-frozen and stored at −80 °C.
Crystallization, Data Collection, and Processing
The protein was crystallized using the hanging-drop vapor-diffusion method at 18 °C. The crystallization conditions for unliganded KpOHCU decarboxylase consisted of either 20% w/v PEG-3000 in Tris-HCl, pH 7.0, with 0.2 m calcium acetate or 22–26% w/v PEG-8000 in sodium cacodylate, pH 6.5, with 0.25 m sodium acetate. Before data collection, the crystals were cryoprotected in the above solutions supplemented with 15–20% ethylene glycol. For the KpOHCU decarboxylase-allantoin complex, crystals were soaked for 5 min in cryosolution that contained 10 mm allantoin. For the allopurinol-treated structures, the crystals were either soaked directly in cryosolution containing 10 mm allopurinol or first treated with cryosolution containing 10 mm allantoin for 5 min and then transferred to cryosolution containing 10 mm allopurinol and removed for data collection after either 1 or 10 min. The crystals of the unliganded protein belonged to space groups P41 (rod-shaped crystals) and P1 (plate-like crystals). After soaking the latter crystals with allantoin or allopurinol, the space group changed to C2. Attempts to soak ligands into the tetragonal crystals caused considerable crystal damage and yielded unusable data.
Data were collected at the Northeastern Collaborative Access Team 24-ID-E beamline of the Advanced Photon Source of Argonne National Laboratory at a wavelength of 0.979 Å and at 0.978 Å at the Cornell High Energy Synchrotron Source. In all cases the data were indexed, integrated, and scaled using HKL-2000 (13). The data collection statistics are summarized in Table 1.
TABLE 1.
Data collection statistics
Numbers in parentheses correspond to the highest resolution shell.
| KpOHCU decarboxylase (P1) | KpOHCU decarboxylase (P41) | KpOHCU decarboxylase-allantoin | KpOHCU decarboxylase-allopurinol soak | |
|---|---|---|---|---|
| Resolution (Å) | 1.79 | 1.55 | 2.00 | 1.98 |
| Wavelength (Å) | 0.987 | 0.987 | 0.987 | 0.987 |
| Beam line | CHESS A1 | CHESS A1 | CHESS A1 | CHESS A1 |
| Space group | P1 | P41 | C2 | C2 |
| a (Å) | 46.8 | 88.2 | 91.2 | 91.4 |
| b (Å) | 49.3 | 47.3 | 46.7 | |
| c (Å) | 51.7 | 40.2 | 50.2 | 47.3 |
| α (°) | 113.2 | |||
| β (°) | 116.5 | 123.1 | 119.0 | |
| γ (°) | 94.3 | |||
| No. of reflections | 54,203 | 367,405 | 29,217 | 27,333 |
| Unique reflections | 31,805 | 49,837 | 12,101 | 11,639 |
| Average I/σ | 27.8 (4.7) | 28.7 (7.3) | 16.5 (2.6) | 17.3 (1.9) |
| Redundancy | 1.7 (1.5) | 7.4 (7.2) | 2.4 (2.0) | 2.3 (1.8) |
| Completeness (%) | 93.8 (75.0) | 99.9 (100.0) | 98.6 (94.1) | 94.7 (58.0) |
| Rsyma (%) | 3.0 (14.2) | 9.6 (37.0) | 7.6 (26.3) | 6.9 (32.6) |
a Rsym = ΣΣi|Ii − 〈I〉|, where 〈I〉 is the mean intensity of the N reflections with intensities Ii and common indices h, k, and l.
Structure Solution and Refinement
All initial attempts at molecular replacement using Phaser (14), MOLREP (15), or CNS (16) with various forms of the A. thaliana OHCU decarboxylase (PDB identifier 2Q37) or the OHCU decarboxylase from zebrafish (PDB identifier 2O70) failed to produce a solution for the higher resolution tetragonal data. Using the P1 data, however, a molecular replacement solution was found using Phaser with an all-alanine search model derived from 2Q37. The model was refined through successive rounds of manual model building using COOT (17) and restrained refinement using REFMAC5 (18). Water molecules were added only after the model converged and was followed by two additional rounds of refinement. The solution of the P1 crystal form was used as a model for molecular replacement for the other data sets. Refinement of the initial model was carried out as detailed above. For the KpOHCU decarboxylase-allantoin complex structure, the ligand position was added directly to the difference Fourier map after the initial rounds of refinement. For this structure, water molecules were added only after the ligand was placed. Refinement statistics are summarized in Table 2.
TABLE 2.
Data refinement statistics
| KpOHCU decarboxylase (P1)-unliganded | KpOHCU decarboxylase (P41)-unliganded | KpOHCU decarboxylase-allantoin | KpOHCU decarboxylase-allopurinol soak | |
|---|---|---|---|---|
| Resolution (Å) | 1.79 | 1.55 | 2.00 | 1.98 |
| No. of protein atoms | 2,528 | 2,305 | 1,270 | 1,065 |
| No. of ligand atoms | 0 | 0 | 11 | 0 |
| No. of water atoms | 246 | 657 | 90 | 78 |
| No. of reflections in working set | 30,202 | 47,280 | 11,525 | 11,067 |
| No. of reflections in test set | 1603 | 3493 | 576 | 554 |
| R factora | 22.1 | 20.5 | 21.3 | 22.3 |
| Rfreeb | 24.5 | 22.9 | 24.2 | 25.8 |
| r.m.s.d. bonds (Å) | 0.006 | 0.005 | 0.006 | 0.006 |
| r.m.s.d. angles (°) | 0.894 | 0.824 | 0.991 | 0.941 |
| Mean B factor (Å2) | 20.1 | 10.8 | 25.1 | 34.2 |
| Coordinate error (Å)c | 0.266 | 0.179 | 0.281 | 0.282 |
| Ramachandran plot | ||||
| Most favored (%) | 96.3 | 97.3 | 95.9 | 95.1 |
| Additionally allowed (%) | 3.7 | 2.7 | 4.1 | 4.9 |
| Generously allowed (%) | 0.0 | 0 | 0 | 0 |
| Disallowed (%) | 0 | 0 | 0 | 0 |
a R factor = Σhkl‖Fobs| − k|Fcalc‖/Σhkl|Fobs|, where Fobs and Fcalc are the observed and calculated structure factors, respectively.
b For Rfree, the sum is extended over a subset of reflections (5%) excluded from all stages of refinement.
c The estimated coordinate error is from the Lazutti plot as calculated by the SFCHECK program (28).
Substrate and Product Modeling
The modeling of the ligands in the active site of KpOHCU decarboxylase was performed using Version 9.7.211 of MacroModel (19, 20). The protein was truncated to a shell containing all atoms within 20 Å of the ligand. Water molecules were removed, and the protein preparation utility was used to add hydrogen atoms and to ensure proper ionic states of amino acid side chains. To model the substrate, OHCU was positioned in the active site by superposition to the allantoin from the KpOHCU decarboxylase-allantoin complex structure. A conformational search of OHCU was used to generate starting conformers for the calculations. For the modeling runs, both the ligand and active site residues were allowed freedom to move. To model the products of the reaction in the active site, the minimized OHCU molecule was used as a starting point. The calculations were completed using the AMBER* force field (21) with a distance dependent dielectric that was further attenuated by a factor of 4. The energy minimization relied upon the TNCG (truncated Newton conjugate gradient) technique (22) and was considered to have converged when the energy gradient was less than 0.01 kJ/mol.
All of the figures presented herein were prepared using PyMOL (23) and ChemBioDraw (CambridgeSoft). The structures have been deposited in the Protein Data Bank with identifiers 3O7H (P1-unliganded), 3O7I (P41-unliganded), 3O7J (C2-allantoin complex), and 3O7K (C2-allopurinol soak).
Steady State Kinetics Measurements
The substrate was generated in situ by treating a urate solution with uricase and HIU hydrolase enzymes. The OHCU was made just before injection by incubating 140 μm urate in 500 μl of 50 mm phosphate buffer, pH 8.0, with 5 μl of a 5 mg/ml uricase solution and 1 μl of a 5-mg/ml HIU hydrolase solution (HpxT from K. pneumoniae). This amount of enzyme was sufficient to completely convert the urate into OHCU in less than 1 min (monitored by the disappearance of the absorbance peak at 300 nm (4)). The decarboxylation reaction was initiated by the addition of 20 μl of 2.5 μm KpOHCU decarboxylase (0.024 nmol). All reactions were carried out at pH 8.0 in 50 mm phosphate buffer in 500 μl total volume. The rate of reaction was monitored by observing the change of absorbance at 256 nm. The initial rate was calculated from the first 15 s after the addition of the enzyme, at which point the reaction had not proceeded beyond 10% toward completion. Duplicate reactions were carried out at each of the various OHCU concentrations. The initial rate data were plotted and fit to the Michaelis-Menten equation using the Kaleidagraph software package (Synergy Software).
Inhibition by Allopurinol
Reaction rates for inhibition studies were measured as described above. To various concentrations of OHCU (generated in situ) was added a solution of 2.5 μm KpOHCU decarboxylase containing allopurinol. The initial rate of reaction, calculated from the first 15 s after the addition of enzyme, was recorded for the various substrate concentrations. The reaction rates were measured at concentrations of 150, 350, and 600 μm allopurinol. A double reciprocal plot was constructed (Fig. 3C), and linear fits to the data were extrapolated to the y intercept to determine the Ki. The reported Ki is the average of the 3 calculated values from the fits to the data at 150, 350, and 600 μm allopurinol.
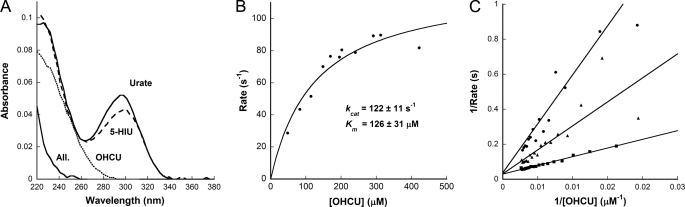
FIGURE 3.
Kinetics of KpOHCU decarboxylase reaction. A, UV traces of the intermediates along the uric acid degradation pathway are shown. Urate (top solid line) is converted to 5-HIU (dashed line) by urate oxidase (HpxO in K. pneumoniae), 5-HIU is converted to OHCU (dotted line) by an HIU hydrolase (HpxT in K. pneumoniae), and OHCU is converted to allantoin (All. lower solid line) by KpOHCU decarboxylase. B, shown are the steady state kinetics of KpOHCU decarboxylase. The rate of OHCU decarboxylation was measured by monitoring the decrease in absorbance at 256 nm. The reactions were followed using a stopped flow apparatus, and the rates were calculated from the first 10 s of the reaction (less than 10% completion). C, shown is a double reciprocal plot showing the competitive inhibition of KpOHCU decarboxylase by allopurinol. The rate was measured at 600 μm inhibitor (filled circles), 350 μm inhibitor (filled triangles), and 150 μm inhibitor (filled squares). The average value of Ki from the three datasets is 31 ± 2 μm.
Circular Dichroism Measurements
The KpOHCU decarboxylase protein was buffer exchanged into a solution of 10 mm phosphate buffer, pH 7.6, and used at a final concentration of 0.1 mg/ml. Data were collected separately for the protein alone, with 1 mm allantoin added, and with 1 mm allopurinol added. The CD spectra were collected on an AVIV Biomedical (Lakewood, NJ) CD spectrometer, Model 202-01. The data were collected at 25 °C from 190 to 260 nm with a 1-nm step size and a 1-nm bandwidth. A 1-mm cell was used, and in all cases, the data reported are an average of three measurements after subtraction of the background signal. Samples run without enzyme present showed that the ligands tested did not contribute to the CD signal at the concentrations used. The programs K2D2 and DicroProt (24, 25) were used for prediction of secondary structure content from the collected data.
RESULTS
Overall Structure of KpOHCU Decarboxylase
The OHCU decarboxylase of K. pneumoniae crystallized with two different morphologies and yielded data in three different space groups. The long rod-shaped crystals that grew in PEG-3000 belonged to space group P41 with two molecules per asymmetric unit and a calculated solvent content of 42.5%. Crystals grown in the PEG-8000 condition were plate-like and belonged to space group P1. This crystal form had two molecules per asymmetric unit with a calculated solvent content of 45.3%. When these crystals were soaked with a solution of mother liquor containing 10 mm allantoin, the space group changed to C2 with one molecule per asymmetric unit, and the solvent content decreased to 43.6%.
KpOHCU decarboxylase is an all-α-helical protein that belongs to the newly discovered OHCU decarboxylase fold (as classified by Pfam). It consists of nine helices in two domains, with the N-terminal domain containing the first four helices and the C terminus of helix 9. Taken together, helices 2 through 4 and helix 9 are consistent with a four-helix bundle. The core of this domain is lined with hydrophobic side chains including two tryptophans, two phenylalanines, and several leucine and valine residues. There are no disulfides in this enzyme, and the fold is stabilized primarily by hydrophobic interactions within the two all α-helical domains.
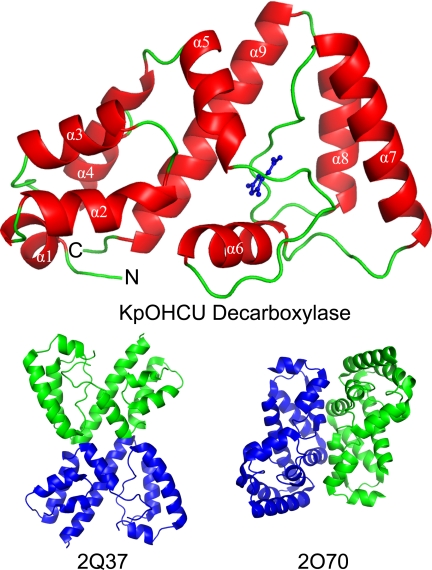
The crystal structures of KpOHCU decarboxylase indicate that this enzyme is a monomer in solution, a conclusion supported by size exclusion chromatography (supplemental Fig. S5). In addition, submission of the structure to the EBI-PISA (26) server, which examines protein interfaces, surfaces, and assemblies, predicted that higher order oligomerization was not likely to be observed. Further evidence comes from the observation that the tetragonal and triclinic/monoclinic crystal forms display completely different packing (Fig. 1 and supplemental Fig. S4). The monomeric structure of KpOHCU decarboxylase with the bound product, allantoin, is shown in Fig. 1A.
FIGURE 1.
Structure of KpOHCU decarboxylase monomer (red helices and green loops) with allantoin bound (blue). The helices are labeled in white text, and the N and C termini are labeled with an N and C, respectively. Also shown are the proposed OHCU decarboxylase dimers from A. thaliana (PDB code 2Q37) and zebrafish (PDB code 2O70).
A search for structurally similar proteins using the DALI server (27) yielded only three homologous structures (supplemental Table S1). Two of these, PDB codes 2Q37 and 2O70, are the OHCU decarboxylase enzymes from A. thaliana (29% sequence identity to HpxQ with a r.m.s.d. of backbone atoms of 1.8) and zebrafish (Danio rerio, 22% sequence identity to HpxQ with an r.m.s.d. of 2.4), respectively (Fig. 1). The third, PDB code 2O8I, is an unpublished structure from Agrobacterium tumefaciens (23% sequence identity to HpxQ with an r.m.s.d. of 2.8) with an uncharacterized function. Based upon the degree of similarity to 2Q37, 2O70, and KpOHCU decarboxylase, it is likely that this protein is also an OHCU decarboxylase.
KpOHCU Decarboxylase Active Site
KpOHCU decarboxylase crystals soaked in mother liquor supplemented with 10 mm allantoin yielded maps with clear difference density in the active site. Although a racemic mixture of both (R)- and (S)-allantoin enantiomers was used for soaking, the density observed in the active site corresponded to a planar molecule, consistent with the enol form of allantoin. The density allowed us to unambiguously place the allantoin molecule in the active site. This placement superimposed well with that of the other OHCU decarboxylase structures.
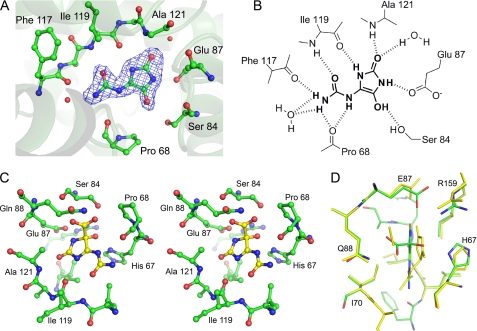
KpOHCU decarboxylase contains one active site per monomer, and all of the contacts with the ligand are made by a single monomer. The active site is formed primarily by the loops connecting helices 5 and 6 and helices 7 and 8. Upon binding, KpOHCU decarboxylase forms a significant number of hydrogen bonds to the ligand (Fig. 2). At least eight non-covalent interactions can be seen between the protein and the ligand, with additional bonding occurring through two active site water molecules that are well ordered in the structure. Most of the polar contacts are with backbone atoms of Pro-68, Phe-117, Ile-199, and Ala-121 with Ser-84 and Glu-87 providing addition bonding through their side chains.
FIGURE 2.
Active site of KpOHCU decarboxylase and ligand contacts. A, shown is the active site with bound product, allantoin, showing Fo − Fc electron density contoured at 3σ. The difference density was calculated before adding the ligand to the model. B, a schematic of ligand binding in KpOHCU decarboxylase active site shows hydrogen bonding between the enzyme and the ligand. C, shown is a stereoview of KpOHCU decarboxylase active site with modeled OHCU (yellow). D, superposition of the energy minimized active site with bound substrate, OHCU (green carbon atoms, red oxygen atoms, blue nitrogen atoms), and the energy minimized active site with bound products, (S)-allantoin and CO2 (yellow) is shown.
Analysis of the B-factors of the crystal structure of both the unliganded and liganded form of KpOHCU decarboxylase shows a relatively even distribution of low B-factors (15 to 20) over the whole protein. The one exception is the helix that spans residues 79–88. Supplemental Fig. S1 shows a ribbon diagram of the KpOHCU decarboxylase structure colored by B-factors. This helix and the adjoining loop regions were examined further both structurally and by CD (see below).
A sequence alignment of KpOHCU decarboxylase with the zebrafish and A. thaliana OHCU decarboxylases is shown in supplemental Fig. S2. Despite the relatively low level of overall similarity between the sequences, there are several conserved regions. In particular, all but one of the active site residues that make polar contacts with the ligand are conserved among the three enzymes (supplemental Fig. S2, blue arrows). The exception is Ala-74 from the A. thaliana OHCU decarboxylase (Ser-84 in KpOHCU decarboxylase). A comparison of the different structures at this residue is impossible because Ala-74 does not appear in the A. thaliana structure as it is part of a presumably disordered loop region.
An alignment of the unliganded and allantoin-bound KpOHCU decarboxylase structures reveals a slight difference in the loop between helices 5 and 6. Although several residues change position, the shift of this region most notably alters the position of His-67, presumably to accommodate the ligand. The superposition of these two structures, illustrating the motion of His-67, is shown in supplemental Fig. S3A.
Modeling of Substrate and Products
To investigate the binding of the substrate and products of the KpOHCU decarboxylase catalyzed reaction, molecular mechanics simulations of OHCU and allantoin/carboxylate in the active site were carried out. Using the structure of the KpOHCU decarboxylase-allantoin complex as a starting point, these molecules were docked into the active site and then energy-minimized using the AMBER* (21) force field. Both the molecules themselves and the active site side chains were allowed freedom of movement. Although most of the residues remain unmoved, the putative active site base, His-67, can adopt several low energy states (supplemental Fig. S3, B and C). Modeling of OHCU also reveals additional hydrogen bonds made between the carboxylate group and Gln-88 as well as between the hydroxyl group and His-67. This structure also shows a shift in the position of the allantoin backbone away from Gln-88 and toward His-67.
Kinetics of KpOHCU Decarboxylase-catalyzed Reaction
The kinetics of conversion of OHCU to allantoin by KpOHCU decarboxylase can be followed spectrophotometrically. Fig. 3A shows the spectra of uric acid overlaid with the degradation products produced during enzymatic conversion to allantoin. The large difference in absorbance of OHCU and allantoin in the region of 240–260 nm allows for the facile measurement of this conversion. By monitoring the change in absorbance at 256 nm, the initial rates of decarboxylation catalyzed by KpOHCU decarboxylase at various OHCU concentrations were measured and used to calculate the steady state parameters (Fig. 3B). The calculated background rate of non-enzymatic decay of OHCU (1.5 ± 0.1 × 10−3 s−1), which was in agreement with the reported value (2.2 × 10−3 s−1 (4)), was accounted for in all cases. The enzyme turns over at a rate of 122 ± 11 s−1 and has a kcat/Km of 9.7 × 105 m−1s−1. An attempt to measure presteady state kinetics of the reaction showed a linear initial rate with no apparent burst phase observed at any point up to the highest detection limit of the apparatus (0.2 ms). In addition, the kinetics were unaffected by viscosity, showing no difference in rate of reaction in the presence of varying concentrations of sucrose or ethylene glycol (data not shown).
Activity of the Q88E Mutant
To examine the role played by Gln-88 in catalysis, the activity of a glutamine to glutamate mutant was measured. Using the identical protocol and conditions as with the native protein, we measured the rate of conversion of OHCU to allantoin by the mutant enzyme. The Q88E mutant was active but turned over at a slower pace than the native enzyme. The maximal rate of this mutant (2.8 ± 0.2 s−1) was 43-fold slower than that of the native rate, whereas the measured Km for the mutant was similar to that of the native enzyme (151 ± 36 μm). The kcat/Km for the mutant was calculated to be 1.9 × 104 m−1s−1.
Inhibition of KpOHCU Decarboxylase by Allopurinol
As there are currently no OHCU decarboxylase inhibitors reported in the literature, we tested several substrate analogues for their ability to inhibit KpOHCU decarboxylase activity. The xanthine oxidase inhibitor, allopurinol (29), was found to reduce the rate of KpOHCU decarboxylase turnover at micromolar concentrations. This compound was determined to be a competitive inhibitor as demonstrated by a double reciprocal plot of inhibition at 150, 350, and 600 μm (Fig. 3C), with a calculated Ki of 30 ± 2 μm.
Structural Observations and CD Measurements of Domain Movement
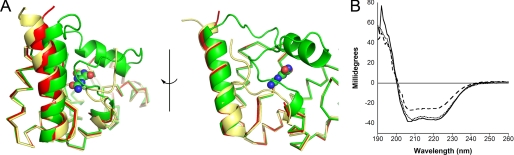
To probe the mechanism of inhibition, we attempted to structurally characterize a complex of KpOHCU decarboxylase and allopurinol. Soaks of the tetragonal crystal form with allopurinol solutions caused crystal degradation and failed to yield usable data. Similar treatment of the monoclinic crystals yielded high quality data with increased symmetry (P1 to C2). Examination of the structure revealed that helix 6 was missing from the density, and the loop regions connecting this helix to the rest of the structure were displaced from the allantoin-bound structure (Fig. 4A).
FIGURE 4.
Conformational change of KpOHCU decarboxylase. A, ribbon diagrams show the structural superposition of KpOHCU decarboxylase-allantoin complex (green), unliganded KpOHCU decarboxylase in tetragonal form (yellow), and KpOHCU decarboxylase treated with allopurinol (red). The r.m.s.d. for the superposition of the allantoin-bound structure to the unliganded is 0.499, whereas the r.m.s.d. for the allopurinol-soaked structure to the unliganded is 0.346. Allantoin is shown in space-fill representation. Helix 6 (above allantoin) is visible only in the allantoin bound structure. B, circular dichroism measurements are shown of unliganded KpOHCU decarboxylase (dotted line), KpOHCU decarboxylase treated with allantoin (solid line), and KpOHCU decarboxylase treated with allopurinol (dashed line). The increase in signal at 208 and 222 nm and the decrease in signal at 193 nm in the presence of allopurinol are all consistent with a loss of helical content.
To examine the possibility that the inhibitor disrupts the tertiary structure of the enzyme, we examined the CD spectra of KpOHCU decarboxylase in the presence and absence of allantoin or allopurinol. The spectra of the protein alone in the presence of allantoin and in the presence of allopurinol are shown in Fig. 4B. Although the presence of allantoin has only a minor influence on the structure, a marked decrease in helical content is apparent in the presence of allopurinol. The approximate helical content drops from 93% in the unliganded structure to 82% in the presence of allopurinol (95% in the presence of allantoin).
DISCUSSION
Despite the relatively low sequence similarity to the most structurally similar model (29% sequence identity to 2Q37), the structure of KpOHCU decarboxylase was solved by molecular replacement. This structure represents the first reported prokaryotic OHCU decarboxylase. Although not a highly similar in sequence, KpOHCU decarboxylase bears a high degree of structural homology to the previously reported OHCU decarboxylases. There are, however, marked distinctions between these enzymes. The most striking of these is the disparity in the quaternary structures.
Although both of the eukaryotic OHCU decarboxylases reported dimers as the biological unit, the spatial relationship and interactions between the two protomers are significantly different (Fig. 1). The K. pneumoniae enzyme presented herein, however, is clearly a monomer in solution. This conclusion is supported by the get filtration data (supplemental Fig. S5), results from the PISA server, and the observation of two different crystal forms with entirely different contacts between monomers in the two structures. Considering that all of the protein-ligand interactions are made by a single monomer, these results suggest that the minimal functional unit of OHCU decarboxylase is a monomer. The dimeric forms present in the eukaryotic structures may be a result of evolutionary pressure. Further biochemical comparison of these enzymes is necessary to determine whether the dimers are more catalytically efficient or are necessary for some other purpose such as protein-protein complex formation.
The active site residues of OHCU decarboxylases are highly conserved (supplemental Fig. S2). Despite this similarity and the observation that this enzyme produces (S)-allantoin stereospecifically, both the (S) and (R) isomers of allantoin have been observed bound in the active site (6, 7). In the KpOHCU decarboxylase structure, however, we observed the enol form of allantoin (Fig. 2). Allantoin is known to undergo slow non-enzymatic racemization, with one likely mechanism for this chemistry being the keto-enol tautomerization (30). In the presence of KpOHCU decarboxylase, this reaction is likely facilitated by His-67, which is well positioned to abstract a proton from allantoin. It is not surprising to see that the enol is stabilized by the active site, as it is structurally more similar to the substrate than either the (R) or (S) enantiomers. Thus, the structure of KpOHCU decarboxylase in complex with this isomer of allantoin allows for a clearer picture of how the substrate interacts with this enzyme and provides a snapshot of an important intermediate in the reaction mechanism.
Positioning of OHCU in the KpOHCU decarboxylase active site is facilitated by the degree of similarity between the substrate and the enol observed in the structure. Although the stereochemistry of enzymatically produced OHCU has not been well characterized, it can be inferred from structures of urate oxidase with bound ligands. The addition of the hydroxyl group to urate occurs at the re- face leading to the (S) isomer of OHCU after hydrolysis by HIU hydrolase cleaves the N1-C6 bond (31).4 A stereo diagram showing the positioning of OHCU in the KpOHCU decarboxylase active site is provided in Fig. 2B. In addition to the hydrogen bonding contacts shown in Fig. 2A, the substrate makes additional bonds to Gln-88 through the carboxylate and with His-67 through the hydroxyl moiety.
To further our understanding of the OHCU decarboxylation reaction, we investigated the kinetics of KpOHCU decarboxylase. To date there have been no reports on the kinetics of a native OHCU decarboxylase. This is likely due to the difficulty of working with the unstable substrate of the reaction. OHCU can be generated in situ by the consecutive reactions of urate oxidase and HIU hydrolase on uric acid and 5-HIU, respectively. The conversion from uric acid to OHCU through 5-HIU can be monitored by UV (Fig. 3A). The clear difference in UV spectra between OHCU and allantoin in the 240–260-nm range allows for the facile measurement of reaction rates. The confounding factor of non-enzymatic decay of 5-HIU and OHCU is negligible if reaction times are kept short. Fig. 3B shows the steady-state kinetics of the KpOHCU decarboxylase-catalyzed decarboxylation reaction and the calculated kinetic parameters. Even though a cofactor is not required, this enzyme is surprisingly efficient with a kcat/Kmof 9.7 × 105 m−1s−1.
Despite the commercial availability of purine derivatives, a search of the literature reveals a paucity of information for small molecule modulators of OHCU decarboxylase activity. There is, however, precedent for binding of such molecules apparent in the crystal structure of zebrafish OHCU decarboxylase in complex with guanine (6). Thus, we began our screen for small molecule modulators of KpOHCU decarboxylase activity with purine analogs. Comparison of wild type KpOHCU decarboxylase activity with activity levels in the presence of these molecules identified allopurinol, a known xanthine oxidase inhibitor. We characterized the inhibition and determined that this molecule is a competitive inhibitor of KpOHCU decarboxylase activity with a Ki of 30 μm.
To probe the interactions between allopurinol and KpOHCU decarboxylase, we first attempted to solve the structure of these two molecules in complex. Our efforts to trap the allopurinol-KpOHCU decarboxylase complex revealed some very interesting details about the conformational flexibility of the enzyme and the mechanism of inhibition. Our first evidence that KpOHCU decarboxylase could exist in different conformational states in solution came from the different structures of the two unliganded crystal forms. The structure of the tetragonal form revealed a non-functional conformation where the crystal packing interactions occurred near the active site (supplemental Fig. S4). Additionally, soaks of the unliganded or liganded KpOHCU decarboxylase crystals with allopurinol showed a similar unfolded state as was observed for the tetragonal form. A superposition of these different conformational states is shown in Fig. 4.
These observations suggested that a region of this enzyme containing helix 6 and bound by the adjoining loop regions was conformationally flexible and was sensitive to active site-ligand interactions. An examination of the B-factors in the full, unliganded, structure suggests that this region of the enzyme has the most dynamic mobility (supplemental Fig. S1). For further evidence of the impact of ligand binding on the conformational state of the enzyme, we measured the circular dichroism spectra of the enzyme in the absence and presence of allantoin and allopurinol. The results, shown in Fig. 4B, indicate that the enzyme has a lower helical content in the presence of the inhibitor. This data taken together with the structural findings indicate that the protein-ligand interactions drive conformational change and that binding of the inhibitor prevents the correct organization of the active site.
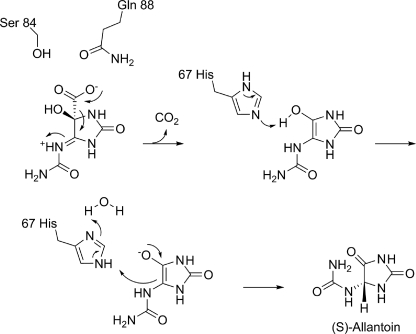
The structural, biochemical, and modeling studies presented herein provide several much needed insights into the mechanism of decarboxylation catalyzed by OHCU decarboxylase enzymes. The proposed mechanism is shown in Scheme 2. KpOHCU decarboxylase is a highly efficient enzyme that operates in a cofactor-independent manner. It has been proposed that OHCU decarboxylase may use a catalytic strategy similar to OMPDC, a widely studied cofactor-independent decarboxylase (6). Although several mechanisms for the decarboxylation reaction catalyzed by OMPDC have been put forth, the mechanism that is most consistent with structural data and quantum mechanical calculations involves ground-state destabilization of the carboxylate group by a proximal aspartate in the active site (11, 12, 32).
SCHEME 2.
Mechanism of OHCU decarboxylation catalyzed by KpOHCU decarboxylase.
Although the active site of KpOHCU decarboxylase contains a large number of hydrogen bonding contacts, locking the substrate into place (as is the case for OMPDC), our data suggest a different mechanism. In the OMPDC decarboxylation reaction, the leaving carboxylate group is destabilized by an unfavorable interaction with an active site aspartate. Based upon our structure, which allows for an accurate placement of the substrate in the active site, the carboxylate group of OHCU will be in close proximity to Gln-88 (Fig. 2, C and D). To examine if this residue provides a destabilizing driving force for decarboxylation, we constructed and measured the rate of reaction of the Q88E mutant. If the KpOHCU decarboxylase catalyzed reaction proceeded in an analogous fashion to that of OMPDC, we would expect a rate increase for the mutant enzyme. What we observed, however, was a decrease in enzyme activity by >40-fold. In addition, the Km of the mutant (151 μm) was similar to that of the native enzyme (126 μm), suggesting that the mutation does not have a large effect upon substrate binding.
Based on this observation, we propose that Gln-88 instead acts to inductively destabilize the carbon-carbon bond between the carboxylate and C5 of the hydantoin ring in a manner analogous to purine nucleoside phosphorylase (33) and related enzymes (34). In the purine nucleoside phosphorylase-catalyzed reaction, an active site asparagine withdraws electrons from the purine ring through hydrogen bond formation causing a weakening and concomitant heterolysis of the C1′-N9 bond. This type of mechanism is consistent with the positioning of Gln-88 in the KpOHCU decarboxylase structure and with the Q88E mutant activity measurements.
The substrate is likely protonated at N6 upon entering the active site (Scheme 2). This would provide an additional driving force for the decarboxylation reaction. A hydrogen bond observed between N6 and the carbonyl oxygen of Pro-68 supports this hypothesis (Fig. 2B).
Upon decarboxylation, an enol intermediate is formed that resembles the isomer seen in our crystal structure (Fig. 2). Modeling of the reaction intermediates suggest that unfavorable interactions between the liberated CO2 and both the enzyme and intermediate force the hydantoin ring toward His-67 (supplemental Fig. S3C). This histidine is ideally positioned to facilitate proton transfer between the hydroxy and C5 of the hydantoin ring. The structures of liganded and unliganded KpOHCU decarboxylase illustrate the conformational flexibility of this residue (supplemental Fig. S3A), and our modeling studies support the role of His-67 in proton transfer (supplemental Fig. S3, B and C). In addition, examination of the H67N mutant in the zebrafish OHCU decarboxylase showed complete loss of activity (6), consistent with its significance for proton transfer.
The enzyme mediated tautomerization illustrated in Scheme 2 is shown to occur in a stepwise fashion. Although it is possible that this transfer could occur by a concerted mechanism, the acidity of the enol suggests that full proton transfer to the histidine followed by ring protonation at C5 could occur as separate steps. This type of charged enolate intermediate is a commonly cited species in several types of enzyme catalyzed reactions (35–39). The tautomerization reaction, facilitated by His-67, yields the product, allantoin, exclusively as the (S) enantiomer. Presteady state kinetic analysis and examination of the effect of viscosity on reaction rate suggest that this latter reaction occurs very rapidly. In addition, as there is no apparent alternative for CO2 escape from the active site, it is likely that the unfavorable interactions caused by the presence of this molecule drive product release.
In summary, we have presented a structural and biochemical characterization of KpOHCU decarboxylase, identified a novel inhibitor, and proposed a mechanism of action for this inhibitor. CD measurements were used to support the structural evidence for dynamic instability of helix 6 and the ability for ligands to modulate this conformational change. The structure of KpOHCU decarboxylase in complex with the enol form of allantoin provides a view of the proposed reaction intermediate and together with the biochemical data and modeling studies allows us to provide a mechanistic analysis of the decarboxylation reaction.
Supplementary Material
Acknowledgments
We thank the staff at the Cornell High Energy Synchrotron Source (CHESS) and Northeastern Collaborative Access Team for advice with data collection and processing. The data collection performed at the CHESS was supported by the National Science Foundation and the NIGMS, National Institutes of Health (NIH) under National Science Foundation Award DMR-0225180 using the Macromolecular Diffraction at the CHESS (MacCHESS) facility, which is supported by National Center for Research Resources, NIH Award RR-01646. Data collected at the Northeastern Collaborative Access Team beamlines of the Advanced Photon Source are supported by National Center for Research Resources, National Institutes of Health Award RR-15301. Use of the Advanced Photon Source is supported by the United States Department of Energy, Office of Basic Energy Sciences under Contract DE-AC02-06CH11357. We acknowledge Cynthia Kinsland of the Protein Production Facility in the Department of Chemistry and Chemical Biology for help with molecular biology and Leslie Kinsland for help with manuscript preparation. We thank M. V. Airola and K. O. Gee of the B. R. Crane laboratory for assistance with the CD data collection.
This work was supported, in whole or in part, by National Institutes of Health Grant GM73220.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S5.
The atomic coordinates and structure factors (codes 3O7H, 3O7I, 3O7J, and 3O7K) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The structures of K. pneumoniae urate oxidase (HpxO) with bound ligands also indicate that the (S) enantiomer of OHCU is produced upon stereospecific oxidation of uric acid by this enzyme (K. A. Hicks and S. E. Ealick, unpublished information).
- HIU
- 5-hydroxyisourate
- OHCU
- 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline
- OMPDC
- orotodine monophosphate decarboxylase
- KpOHCU decarboxylase
- K. pneumoniae OHCU decarboxylase
- r.m.s.d
- root mean square deviation
- CHESS
- Cornell High Energy Synchrotron Source.
REFERENCES
- 1.Vogels G. D., Van der Drift C. (1976) Bacteriol. Rev. 40, 403–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zrenner R., Stitt M., Sonnewald U., Boldt R. (2006) Annu. Rev. Plant Biol. 57, 805–836 [DOI] [PubMed] [Google Scholar]
- 3.Kahn K., Tipton P. A. (1997) Biochemistry 36, 4731–4738 [DOI] [PubMed] [Google Scholar]
- 4.Kahn K., Tipton P. A. (1998) Biochemistry 37, 11651–11659 [DOI] [PubMed] [Google Scholar]
- 5.Ramazzina I., Folli C., Secchi A., Berni R., Percudani R. (2006) Nat. Chem. Biol. 2, 144–148 [DOI] [PubMed] [Google Scholar]
- 6.Cendron L., Berni R., Folli C., Ramazzina I., Percudani R., Zanotti G. (2007) J. Biol. Chem. 282, 18182–18189 [DOI] [PubMed] [Google Scholar]
- 7.Kim K., Park J., Rhee S. (2007) J. Biol. Chem. 282, 23457–23464 [DOI] [PubMed] [Google Scholar]
- 8.Frey P. A., Hegeman A. D. (2007) Enzymatic Reaction Mechanisms, pp. 387–390, Oxford University Press, New York [Google Scholar]
- 9.McMurry J. E., Begley T. P. (2005) The Organic Chemistry of Biological Pathways, pp. 134–135, 251, Roberts and Co., Englewood, CO [Google Scholar]
- 10.Appleby T. C., Kinsland C., Begley T. P., Ealick S. E. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 2005–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Begley T. P., Appleby T. C., Ealick S. E. (2000) Curr. Opin. Struct. Biol. 10, 711–718 [DOI] [PubMed] [Google Scholar]
- 12.Wood B. M., Chan K. K., Amyes T. L., Richard J. P., Gerlt J. A. (2009) Biochemistry 48, 5510–5517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otwinowski Z., Minor W. (1997) in Methods in Enzymology (Carter C. W., Jr., Sweet R. M. eds) Vol. 276, pp. 307–326, Academic Press, New York: [DOI] [PubMed] [Google Scholar]
- 14.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vagin A., Teplyakov A. (2000) Acta Crystallogr. D 56, 1622–1624 [DOI] [PubMed] [Google Scholar]
- 16.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 17.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 18.Murshudov G. N., Vagin A. A., Lebedev A., Wilson K. S., Dodson E. J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 247–255 [DOI] [PubMed] [Google Scholar]
- 19.Mohamadi F., Richards N. G. J., Guida W. C., Liskamp R., Lipton M., Caufield C., Chang G., Hendrickson T., Still W. C. (1990) J. Comput. Chem. 11, 440–467 [Google Scholar]
- 20.Still W. C., Mohamadi F., Richards N. G. J., Guida W. C., Liskamp R., Lipton M., Caulfield C., Chang G., Hendrickson T. (1999) MacroModel, 2.0 Ed., Columbia University, New York, NY [Google Scholar]
- 21.Weiner S. J., Kollman P. A., Case D. A., Singh U. C., Ghio C., Alagona G., Profeta S., Jr., Weiner P. (1984) J. Am. Chem. Soc. 106, 765–784 [Google Scholar]
- 22.Ponder J. W., Richards F. M. (1987) J. Comput. Chem. 8, 1016–1024 [Google Scholar]
- 23.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 24.Deleage G., Geourjon C. (1993) Comput. Appl. Biosci. 9, 197–199 [DOI] [PubMed] [Google Scholar]
- 25.Perez-Iratxeta C., Andrade-Navarro M. A. (2008) BMC Struct. Biol. 8, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krissinel E., Henrick K. (2005) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 27.Holm L., Kääriäinen S., Rosenström P., Schenkel A. (2008) Bioinformatics 24, 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaguine A. A., Richelle J., Wodak S. J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 191–205 [DOI] [PubMed] [Google Scholar]
- 29.Borges F., Fernandes E., Roleira F. (2002) Curr. Med. Chem. 9, 195–217 [DOI] [PubMed] [Google Scholar]
- 30.Kahn K., Tipton P. A. (2000) Bioorg. Chem. 28, 62–72 [Google Scholar]
- 31.Gabison L., Prangé T., Colloc'h N., El Hajji M., Castro B., Chiadmi M. (2008) BMC Struct. Biol. 8, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Begley T. P., Ealick S. E. (2004) Curr. Opin. Chem. Biol. 8, 508–515 [DOI] [PubMed] [Google Scholar]
- 33.Fedorov A., Shi W., Kicska G., Fedorov E., Tyler P. C., Furneaux R. H., Hanson J. C., Gainsford G. J., Larese J. Z., Schramm V. L., Almo S. C. (2001) Biochemistry 40, 853–860 [DOI] [PubMed] [Google Scholar]
- 34.Doukov T. I., Hemmi H., Drennan C. L., Ragsdale S. W. (2007) J. Biol. Chem. 282, 6609–6618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heck S. D., Faraci W. S., Kelbaugh P. R., Saccomano N. A., Thadeio P. F., Volkmann R. A. (1996) Proc. Natl. Acad. Sci. 93, 4036–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Imker H. J., Fedorov A. A., Fedorov E. V., Almo S. C., Gerlt J. A. (2007) Biochemistry 46, 4077–4089 [DOI] [PubMed] [Google Scholar]
- 37.Rudik I., Thorpe C. (2001) Arch. Biochem. Biophys. 392, 341–348 [DOI] [PubMed] [Google Scholar]
- 38.Watanabe K., Mie T., Ichihara A., Oikawa H., Honma M. (2000) J. Biol. Chem. 275, 38393–38401 [DOI] [PubMed] [Google Scholar]
- 39.Albery W. J., Knowled J. R. (1986) Biochemistry 25, 2572–2577 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.